Please be patient as the page loads
|
RP9_MOUSE
|
||||||
| SwissProt Accessions | P97762 | Gene names | Rp9, Rp9h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinitis pigmentosa 9 protein homolog (Pim-1-associated protein) (PAP-1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
RP9_MOUSE
|
||||||
| θ value | 4.35321e-87 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97762 | Gene names | Rp9, Rp9h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinitis pigmentosa 9 protein homolog (Pim-1-associated protein) (PAP-1). | |||||
|
RP9_HUMAN
|
||||||
| θ value | 3.22841e-82 (rank : 2) | NC score | 0.975575 (rank : 2) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TA86 | Gene names | RP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinitis pigmentosa 9 protein (Pim-1-associated protein) (PAP-1). | |||||
|
CIR_MOUSE
|
||||||
| θ value | 0.00665767 (rank : 3) | NC score | 0.223611 (rank : 4) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
CIR_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 4) | NC score | 0.225101 (rank : 3) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
STMN1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 5) | NC score | 0.105719 (rank : 5) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 6) | NC score | 0.094322 (rank : 8) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 7) | NC score | 0.093117 (rank : 9) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
STMN1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 8) | NC score | 0.103193 (rank : 6) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
ANR12_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.058062 (rank : 11) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
PLCB4_HUMAN
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.045855 (rank : 12) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15147, Q5JYS8, Q5JYT0, Q5JYT4, Q9BQW5, Q9BQW6, Q9BQW8, Q9UJQ2 | Gene names | PLCB4 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 4 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-4) (PLC-beta-4). | |||||
|
K0831_MOUSE
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.094925 (rank : 7) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CDJ3, Q69ZY1, Q6PFY6, Q8C6N0, Q8R3M3 | Gene names | Kiaa0831, D14Ertd436e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
ZN292_HUMAN
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.036496 (rank : 16) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
KIF2C_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.015268 (rank : 23) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99661, Q96C18, Q96HB8, Q9BWV8 | Gene names | KIF2C, KNSL6 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF2C (Mitotic centromere-associated kinesin) (MCAK) (Kinesin-like protein 6). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.017732 (rank : 20) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
PK1IP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.018817 (rank : 19) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCE5, Q3UBK7, Q80UT4, Q8C5N6, Q923K2, Q9D3E9 | Gene names | Pak1ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p21-activated protein kinase-interacting protein 1 (PAK1-interacting protein 1) (Putative PAK inhibitor Skb15). | |||||
|
RIOK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.042470 (rank : 14) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
K0831_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.090304 (rank : 10) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZNE5, O94920 | Gene names | KIAA0831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
PPIG_HUMAN
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.042121 (rank : 15) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
SEM3D_HUMAN
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.008965 (rank : 24) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95025, Q6UW77, Q8NCQ1 | Gene names | SEMA3D | |||
|
Domain Architecture |
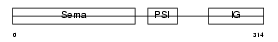
|
|||||
| Description | Semaphorin-3D precursor. | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.043560 (rank : 13) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.015603 (rank : 22) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
DDX10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.006487 (rank : 25) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||
|
INVO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.029872 (rank : 17) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
MDM4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.016371 (rank : 21) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35618 | Gene names | Mdm4, Mdmx | |||
|
Domain Architecture |
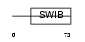
|
|||||
| Description | Mdm4 protein (p53-binding protein Mdm4) (Mdm2-like p53-binding protein) (Mdmx protein) (Double minute 4 protein). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.029526 (rank : 18) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
RP9_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.35321e-87 (rank : 1) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P97762 | Gene names | Rp9, Rp9h | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinitis pigmentosa 9 protein homolog (Pim-1-associated protein) (PAP-1). | |||||
|
RP9_HUMAN
|
||||||
| NC score | 0.975575 (rank : 2) | θ value | 3.22841e-82 (rank : 2) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8TA86 | Gene names | RP9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinitis pigmentosa 9 protein (Pim-1-associated protein) (PAP-1). | |||||
|
CIR_HUMAN
|
||||||
| NC score | 0.225101 (rank : 3) | θ value | 0.00869519 (rank : 4) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
CIR_MOUSE
|
||||||
| NC score | 0.223611 (rank : 4) | θ value | 0.00665767 (rank : 3) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DA19, Q3V2N9, Q4KL44, Q52KL9, Q5FW66 | Gene names | Cir | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor. | |||||
|
STMN1_MOUSE
|
||||||
| NC score | 0.105719 (rank : 5) | θ value | 0.0431538 (rank : 5) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
STMN1_HUMAN
|
||||||
| NC score | 0.103193 (rank : 6) | θ value | 0.0563607 (rank : 8) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
K0831_MOUSE
|
||||||
| NC score | 0.094925 (rank : 7) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CDJ3, Q69ZY1, Q6PFY6, Q8C6N0, Q8R3M3 | Gene names | Kiaa0831, D14Ertd436e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.094322 (rank : 8) | θ value | 0.0431538 (rank : 6) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.093117 (rank : 9) | θ value | 0.0431538 (rank : 7) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
K0831_HUMAN
|
||||||
| NC score | 0.090304 (rank : 10) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6ZNE5, O94920 | Gene names | KIAA0831 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0831. | |||||
|
ANR12_HUMAN
|
||||||
| NC score | 0.058062 (rank : 11) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1081 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q6UB98, O94951, Q658K1, Q6QMF7, Q9H231, Q9H784 | Gene names | ANKRD12, ANCO2, KIAA0874 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 12 (Ankyrin repeat-containing cofactor 2) (GAC-1 protein). | |||||
|
PLCB4_HUMAN
|
||||||
| NC score | 0.045855 (rank : 12) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15147, Q5JYS8, Q5JYT0, Q5JYT4, Q9BQW5, Q9BQW6, Q9BQW8, Q9UJQ2 | Gene names | PLCB4 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 4 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-4) (PLC-beta-4). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.043560 (rank : 13) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
RIOK1_HUMAN
|
||||||
| NC score | 0.042470 (rank : 14) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BRS2, Q8NDC8, Q96NV9 | Gene names | RIOK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase RIO1 (EC 2.7.11.1) (RIO kinase 1). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.042121 (rank : 15) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
ZN292_HUMAN
|
||||||
| NC score | 0.036496 (rank : 16) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
INVO_MOUSE
|
||||||
| NC score | 0.029872 (rank : 17) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 474 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P48997 | Gene names | Ivl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.029526 (rank : 18) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
PK1IP_MOUSE
|
||||||
| NC score | 0.018817 (rank : 19) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DCE5, Q3UBK7, Q80UT4, Q8C5N6, Q923K2, Q9D3E9 | Gene names | Pak1ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p21-activated protein kinase-interacting protein 1 (PAK1-interacting protein 1) (Putative PAK inhibitor Skb15). | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.017732 (rank : 20) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
MDM4_MOUSE
|
||||||
| NC score | 0.016371 (rank : 21) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O35618 | Gene names | Mdm4, Mdmx | |||
|
Domain Architecture |

|
|||||
| Description | Mdm4 protein (p53-binding protein Mdm4) (Mdm2-like p53-binding protein) (Mdmx protein) (Double minute 4 protein). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.015603 (rank : 22) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
KIF2C_HUMAN
|
||||||
| NC score | 0.015268 (rank : 23) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 138 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99661, Q96C18, Q96HB8, Q9BWV8 | Gene names | KIF2C, KNSL6 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like protein KIF2C (Mitotic centromere-associated kinesin) (MCAK) (Kinesin-like protein 6). | |||||
|
SEM3D_HUMAN
|
||||||
| NC score | 0.008965 (rank : 24) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O95025, Q6UW77, Q8NCQ1 | Gene names | SEMA3D | |||
|
Domain Architecture |

|
|||||
| Description | Semaphorin-3D precursor. | |||||
|
DDX10_MOUSE
|
||||||
| NC score | 0.006487 (rank : 25) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 25 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80Y44 | Gene names | Ddx10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX10 (EC 3.6.1.-) (DEAD box protein 10). | |||||