Please be patient as the page loads
|
PHOP2_HUMAN
|
||||||
| SwissProt Accessions | Q8TCD6 | Gene names | PHOSPHO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyridoxal phosphate phosphatase PHOSPHO2 (EC 3.1.3.74). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PHOP2_HUMAN
|
||||||
| θ value | 1.05497e-141 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TCD6 | Gene names | PHOSPHO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyridoxal phosphate phosphatase PHOSPHO2 (EC 3.1.3.74). | |||||
|
PHOP2_MOUSE
|
||||||
| θ value | 4.03687e-117 (rank : 2) | NC score | 0.995149 (rank : 2) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D9M5, Q3V1M5 | Gene names | Phospho2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyridoxal phosphate phosphatase PHOSPHO2 (EC 3.1.3.74). | |||||
|
PHOP1_MOUSE
|
||||||
| θ value | 1.90589e-58 (rank : 3) | NC score | 0.944081 (rank : 3) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R2H9 | Gene names | Phospho1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoethanolamine/phosphocholine phosphatase (EC 3.1.3.75). | |||||
|
PHOP1_HUMAN
|
||||||
| θ value | 4.69435e-57 (rank : 4) | NC score | 0.942391 (rank : 4) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TCT1 | Gene names | PHOSPHO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoethanolamine/phosphocholine phosphatase (EC 3.1.3.75). | |||||
|
RA6I1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 5) | NC score | 0.074037 (rank : 5) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
RA6I1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 6) | NC score | 0.073354 (rank : 6) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
ZN567_HUMAN
|
||||||
| θ value | 0.47712 (rank : 7) | NC score | 0.004521 (rank : 9) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N184, Q6N044 | Gene names | ZNF567 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 567. | |||||
|
ZNF25_HUMAN
|
||||||
| θ value | 0.62314 (rank : 8) | NC score | 0.004288 (rank : 10) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 763 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17030, Q8IYE3, Q8NDD6, Q96MU2 | Gene names | ZNF25, KOX19 | |||
|
Domain Architecture |
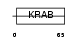
|
|||||
| Description | Zinc finger protein 25 (Zinc finger protein KOX19). | |||||
|
CETN3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 9) | NC score | 0.029092 (rank : 7) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |
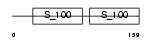
|
|||||
| Description | Centrin-3. | |||||
|
CETN3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 10) | NC score | 0.027082 (rank : 8) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35648 | Gene names | Cetn3, Cen3 | |||
|
Domain Architecture |
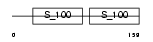
|
|||||
| Description | Centrin-3. | |||||
|
PHOP2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.05497e-141 (rank : 1) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TCD6 | Gene names | PHOSPHO2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyridoxal phosphate phosphatase PHOSPHO2 (EC 3.1.3.74). | |||||
|
PHOP2_MOUSE
|
||||||
| NC score | 0.995149 (rank : 2) | θ value | 4.03687e-117 (rank : 2) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D9M5, Q3V1M5 | Gene names | Phospho2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pyridoxal phosphate phosphatase PHOSPHO2 (EC 3.1.3.74). | |||||
|
PHOP1_MOUSE
|
||||||
| NC score | 0.944081 (rank : 3) | θ value | 1.90589e-58 (rank : 3) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8R2H9 | Gene names | Phospho1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoethanolamine/phosphocholine phosphatase (EC 3.1.3.75). | |||||
|
PHOP1_HUMAN
|
||||||
| NC score | 0.942391 (rank : 4) | θ value | 4.69435e-57 (rank : 4) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8TCT1 | Gene names | PHOSPHO1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphoethanolamine/phosphocholine phosphatase (EC 3.1.3.75). | |||||
|
RA6I1_HUMAN
|
||||||
| NC score | 0.074037 (rank : 5) | θ value | 0.125558 (rank : 5) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6IQ26, Q96GN3, Q9H6U7, Q9UFV0, Q9UPR1 | Gene names | RAB6IP1, KIAA1091 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
RA6I1_MOUSE
|
||||||
| NC score | 0.073354 (rank : 6) | θ value | 0.163984 (rank : 6) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6PAL8, Q62146, Q8C829, Q8VDF6, Q9QYZ2 | Gene names | Rab6ip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab6-interacting protein 1 (Rab6IP1). | |||||
|
CETN3_HUMAN
|
||||||
| NC score | 0.029092 (rank : 7) | θ value | 3.0926 (rank : 9) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15182, Q9BS23 | Gene names | CETN3, CEN3 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-3. | |||||
|
CETN3_MOUSE
|
||||||
| NC score | 0.027082 (rank : 8) | θ value | 3.0926 (rank : 10) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35648 | Gene names | Cetn3, Cen3 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-3. | |||||
|
ZN567_HUMAN
|
||||||
| NC score | 0.004521 (rank : 9) | θ value | 0.47712 (rank : 7) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 769 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8N184, Q6N044 | Gene names | ZNF567 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 567. | |||||
|
ZNF25_HUMAN
|
||||||
| NC score | 0.004288 (rank : 10) | θ value | 0.62314 (rank : 8) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 763 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P17030, Q8IYE3, Q8NDD6, Q96MU2 | Gene names | ZNF25, KOX19 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 25 (Zinc finger protein KOX19). | |||||