Please be patient as the page loads
|
PEX19_MOUSE
|
||||||
| SwissProt Accessions | Q8VCI5, Q8CEE1, Q921H0, Q9CZC1, Q9QUQ1 | Gene names | Pex19, Pxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal biogenesis factor 19 (Peroxin-19) (Peroxisomal farnesylated protein) (PxF). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PEX19_MOUSE
|
||||||
| θ value | 2.59848e-140 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8VCI5, Q8CEE1, Q921H0, Q9CZC1, Q9QUQ1 | Gene names | Pex19, Pxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal biogenesis factor 19 (Peroxin-19) (Peroxisomal farnesylated protein) (PxF). | |||||
|
PEX19_HUMAN
|
||||||
| θ value | 4.30305e-127 (rank : 2) | NC score | 0.983785 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P40855, Q8NI97 | Gene names | PEX19, HK33, PXF | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal biogenesis factor 19 (Peroxin-19) (Peroxisomal farnesylated protein) (33 kDa housekeeping protein). | |||||
|
MRCKG_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 3) | NC score | 0.025704 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
CALD1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 4) | NC score | 0.046546 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
REST_HUMAN
|
||||||
| θ value | 0.163984 (rank : 5) | NC score | 0.037798 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 6) | NC score | 0.038400 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
LIPA3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 7) | NC score | 0.048584 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
CETN2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 8) | NC score | 0.039210 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |
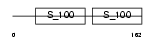
|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
EXOC5_HUMAN
|
||||||
| θ value | 1.38821 (rank : 9) | NC score | 0.083866 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00471 | Gene names | EXOC5, SEC10, SEC10L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 5 (Exocyst complex component Sec10) (hSec10). | |||||
|
EXOC5_MOUSE
|
||||||
| θ value | 1.38821 (rank : 10) | NC score | 0.083636 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3TPX4 | Gene names | Exoc5, Sec10l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 5 (Exocyst complex component Sec10). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 1.81305 (rank : 11) | NC score | 0.035531 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 12) | NC score | 0.032740 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.032433 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 14) | NC score | 0.039716 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
PCQAP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.042538 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
CETN2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.033898 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
GEMI_MOUSE
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.042053 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88513 | Gene names | Gmnn | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
JHD2B_MOUSE
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.019595 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.030935 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
K2C6G_MOUSE
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.020742 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R0H5, Q7TPF3, Q9D0X6 | Gene names | Krt6g, K6irs1, Krt2-6g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 6G (Cytokeratin-6G) (CK 6G) (K6g keratin) (Keratin-K6irs) (mK6irs1/Krt2-6g). | |||||
|
STX7_MOUSE
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.027867 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70439 | Gene names | Stx7, Syn7 | |||
|
Domain Architecture |
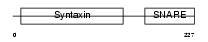
|
|||||
| Description | Syntaxin-7. | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.030467 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.031379 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
BUB1B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 24) | NC score | 0.019853 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60566, O60501, O60627, O60758, O75389, Q96KM4 | Gene names | BUB1B, BUBR1, MAD3L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (hBUBR1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L) (SSK1). | |||||
|
CCNB3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.021860 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
LIPA3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.033516 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60469 | Gene names | Ppfia3 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
NR2E3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.010128 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QXZ7 | Gene names | Nr2e3, Pnr, Rnr | |||
|
Domain Architecture |
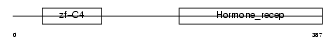
|
|||||
| Description | Photoreceptor-specific nuclear receptor (Retina-specific nuclear receptor). | |||||
|
NRIP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.019697 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CBD1, Q8C3Y8, Q9Z2K2 | Gene names | Nrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
RXRG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.010346 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48443 | Gene names | RXRG, NR2B3 | |||
|
Domain Architecture |
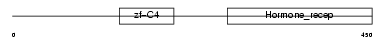
|
|||||
| Description | Retinoic acid receptor RXR-gamma (Retinoid X receptor gamma). | |||||
|
RXRG_MOUSE
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.010263 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28705 | Gene names | Rxrg, Nr2b3 | |||
|
Domain Architecture |
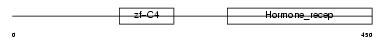
|
|||||
| Description | Retinoic acid receptor RXR-gamma (Retinoid X receptor gamma). | |||||
|
TXLNA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.032133 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
CETN1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.019744 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |
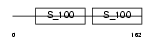
|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.031110 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
HOOK3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.027681 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
KIF4A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.022292 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
STX7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.024644 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15400, Q5SZW2, Q96ES9 | Gene names | STX7 | |||
|
Domain Architecture |
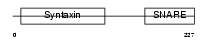
|
|||||
| Description | Syntaxin-7. | |||||
|
TTC5_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.027120 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99LG4 | Gene names | Ttc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 5 (TPR repeat protein 5). | |||||
|
PEX19_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.59848e-140 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8VCI5, Q8CEE1, Q921H0, Q9CZC1, Q9QUQ1 | Gene names | Pex19, Pxf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal biogenesis factor 19 (Peroxin-19) (Peroxisomal farnesylated protein) (PxF). | |||||
|
PEX19_HUMAN
|
||||||
| NC score | 0.983785 (rank : 2) | θ value | 4.30305e-127 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P40855, Q8NI97 | Gene names | PEX19, HK33, PXF | |||
|
Domain Architecture |

|
|||||
| Description | Peroxisomal biogenesis factor 19 (Peroxin-19) (Peroxisomal farnesylated protein) (33 kDa housekeeping protein). | |||||
|
EXOC5_HUMAN
|
||||||
| NC score | 0.083866 (rank : 3) | θ value | 1.38821 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O00471 | Gene names | EXOC5, SEC10, SEC10L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 5 (Exocyst complex component Sec10) (hSec10). | |||||
|
EXOC5_MOUSE
|
||||||
| NC score | 0.083636 (rank : 4) | θ value | 1.38821 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q3TPX4 | Gene names | Exoc5, Sec10l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 5 (Exocyst complex component Sec10). | |||||
|
LIPA3_HUMAN
|
||||||
| NC score | 0.048584 (rank : 5) | θ value | 0.813845 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.046546 (rank : 6) | θ value | 0.125558 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
PCQAP_HUMAN
|
||||||
| NC score | 0.042538 (rank : 7) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 549 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96RN5, O15413, Q6IC31, Q8NF16, Q96CT0, Q96IH7, Q9P1T3 | Gene names | PCQAP, ARC105, CTG7A, TIG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (TPA-inducible gene 1 protein) (TIG-1) (Activator-recruited cofactor 105 kDa component) (ARC105) (CTG repeat protein 7a). | |||||
|
GEMI_MOUSE
|
||||||
| NC score | 0.042053 (rank : 8) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O88513 | Gene names | Gmnn | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.039716 (rank : 9) | θ value | 2.36792 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
CETN2_HUMAN
|
||||||
| NC score | 0.039210 (rank : 10) | θ value | 1.38821 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P41208, Q53XW1 | Gene names | CETN2, CALT, CEN2 | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.038400 (rank : 11) | θ value | 0.47712 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.037798 (rank : 12) | θ value | 0.163984 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.035531 (rank : 13) | θ value | 1.81305 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CETN2_MOUSE
|
||||||
| NC score | 0.033898 (rank : 14) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R1K9, Q3UBB4, Q9CWM0 | Gene names | Cetn2, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-2 (Caltractin isoform 1). | |||||
|
LIPA3_MOUSE
|
||||||
| NC score | 0.033516 (rank : 15) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60469 | Gene names | Ppfia3 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.032740 (rank : 16) | θ value | 1.81305 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.032433 (rank : 17) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
TXLNA_MOUSE
|
||||||
| NC score | 0.032133 (rank : 18) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 846 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6PAM1, Q6P1E5 | Gene names | Txlna, Txln | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-taxilin. | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.031379 (rank : 19) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.031110 (rank : 20) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.030935 (rank : 21) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.030467 (rank : 22) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
STX7_MOUSE
|
||||||
| NC score | 0.027867 (rank : 23) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O70439 | Gene names | Stx7, Syn7 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-7. | |||||
|
HOOK3_MOUSE
|
||||||
| NC score | 0.027681 (rank : 24) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8BUK6, Q8BV48, Q8BW57, Q8BY41 | Gene names | Hook3 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 3. | |||||
|
TTC5_MOUSE
|
||||||
| NC score | 0.027120 (rank : 25) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99LG4 | Gene names | Ttc5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tetratricopeptide repeat protein 5 (TPR repeat protein 5). | |||||
|
MRCKG_HUMAN
|
||||||
| NC score | 0.025704 (rank : 26) | θ value | 0.0148317 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
STX7_HUMAN
|
||||||
| NC score | 0.024644 (rank : 27) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O15400, Q5SZW2, Q96ES9 | Gene names | STX7 | |||
|
Domain Architecture |

|
|||||
| Description | Syntaxin-7. | |||||
|
KIF4A_HUMAN
|
||||||
| NC score | 0.022292 (rank : 28) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O95239, Q86TN3, Q86XX7, Q9NNY6, Q9NY24, Q9UMW3 | Gene names | KIF4A, KIF4 | |||
|
Domain Architecture |

|
|||||
| Description | Chromosome-associated kinesin KIF4A (Chromokinesin). | |||||
|
CCNB3_MOUSE
|
||||||
| NC score | 0.021860 (rank : 29) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q810T2, Q810T3, Q8VDC8 | Gene names | Ccnb3, Cycb3 | |||
|
Domain Architecture |

|
|||||
| Description | G2/mitotic-specific cyclin-B3. | |||||
|
K2C6G_MOUSE
|
||||||
| NC score | 0.020742 (rank : 30) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9R0H5, Q7TPF3, Q9D0X6 | Gene names | Krt6g, K6irs1, Krt2-6g | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cytoskeletal 6G (Cytokeratin-6G) (CK 6G) (K6g keratin) (Keratin-K6irs) (mK6irs1/Krt2-6g). | |||||
|
BUB1B_HUMAN
|
||||||
| NC score | 0.019853 (rank : 31) | θ value | 6.88961 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60566, O60501, O60627, O60758, O75389, Q96KM4 | Gene names | BUB1B, BUBR1, MAD3L | |||
|
Domain Architecture |

|
|||||
| Description | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta (EC 2.7.11.1) (hBUBR1) (MAD3/BUB1-related protein kinase) (Mitotic checkpoint kinase MAD3L) (SSK1). | |||||
|
CETN1_MOUSE
|
||||||
| NC score | 0.019744 (rank : 32) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P41209, Q3V119, Q9D9G9, Q9DAL6 | Gene names | Cetn1, Calt | |||
|
Domain Architecture |

|
|||||
| Description | Centrin-1 (Caltractin). | |||||
|
NRIP1_MOUSE
|
||||||
| NC score | 0.019697 (rank : 33) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8CBD1, Q8C3Y8, Q9Z2K2 | Gene names | Nrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear receptor-interacting protein 1 (Nuclear factor RIP140) (Receptor-interacting protein 140). | |||||
|
JHD2B_MOUSE
|
||||||
| NC score | 0.019595 (rank : 34) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZPY7, Q2VPQ5, Q5U5V7, Q6P9K3, Q8CCE2, Q8K2A5, Q9CU57 | Gene names | Jmjd1b, Jhdm2b, Kiaa1082 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2B (EC 1.14.11.-) (Jumonji domain-containing protein 1B). | |||||
|
RXRG_HUMAN
|
||||||
| NC score | 0.010346 (rank : 35) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P48443 | Gene names | RXRG, NR2B3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-gamma (Retinoid X receptor gamma). | |||||
|
RXRG_MOUSE
|
||||||
| NC score | 0.010263 (rank : 36) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28705 | Gene names | Rxrg, Nr2b3 | |||
|
Domain Architecture |

|
|||||
| Description | Retinoic acid receptor RXR-gamma (Retinoid X receptor gamma). | |||||
|
NR2E3_MOUSE
|
||||||
| NC score | 0.010128 (rank : 37) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QXZ7 | Gene names | Nr2e3, Pnr, Rnr | |||
|
Domain Architecture |

|
|||||
| Description | Photoreceptor-specific nuclear receptor (Retina-specific nuclear receptor). | |||||