Please be patient as the page loads
|
PERL_HUMAN
|
||||||
| SwissProt Accessions | P22079, Q13408 | Gene names | LPO, SAPX | |||
|
Domain Architecture |

|
|||||
| Description | Lactoperoxidase precursor (EC 1.11.1.7) (LPO) (Salivary peroxidase) (SPO). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PERE_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.990515 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11678, Q4TVP3 | Gene names | EPX, EPER, EPO, EPP | |||
|
Domain Architecture |

|
|||||
| Description | Eosinophil peroxidase precursor (EC 1.11.1.7) (EPO) [Contains: Eosinophil peroxidase light chain; Eosinophil peroxidase heavy chain]. | |||||
|
PERE_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.989455 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49290, Q61798 | Gene names | Epx, Eper | |||
|
Domain Architecture |

|
|||||
| Description | Eosinophil peroxidase precursor (EC 1.11.1.7) (EPO) [Contains: Eosinophil peroxidase light chain; Eosinophil peroxidase heavy chain]. | |||||
|
PERL_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P22079, Q13408 | Gene names | LPO, SAPX | |||
|
Domain Architecture |

|
|||||
| Description | Lactoperoxidase precursor (EC 1.11.1.7) (LPO) (Salivary peroxidase) (SPO). | |||||
|
PERM_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.991810 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P05164, Q14862, Q4PJH5 | Gene names | MPO | |||
|
Domain Architecture |

|
|||||
| Description | Myeloperoxidase precursor (EC 1.11.1.7) (MPO) [Contains: 89 kDa myeloperoxidase; 84 kDa myeloperoxidase; Myeloperoxidase light chain; Myeloperoxidase heavy chain]. | |||||
|
PERM_MOUSE
|
||||||
| θ value | 0 (rank : 5) | NC score | 0.992369 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11247 | Gene names | Mpo | |||
|
Domain Architecture |

|
|||||
| Description | Myeloperoxidase precursor (EC 1.11.1.7) (MPO) [Contains: Myeloperoxidase light chain; Myeloperoxidase heavy chain]. | |||||
|
PERT_HUMAN
|
||||||
| θ value | 1.91815e-151 (rank : 6) | NC score | 0.851853 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P07202, P09934, P09935, Q8IUL0, Q8NF94, Q8NF95, Q8NF96, Q8NF97, Q8TCI9 | Gene names | TPO | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
PERT_MOUSE
|
||||||
| θ value | 2.12077e-150 (rank : 7) | NC score | 0.859008 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35419, Q8C8B1 | Gene names | Tpo | |||
|
Domain Architecture |
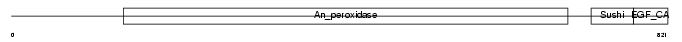
|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
DUOX1_HUMAN
|
||||||
| θ value | 1.73987e-27 (rank : 8) | NC score | 0.514553 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRD9, Q6ZMB3, Q6ZR09, Q9NZC1 | Gene names | DUOX1, DUOX, LNOX1, THOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 1 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH thyroid oxidase 1) (Thyroid oxidase 1) (Large NOX 1) (Long NOX 1). | |||||
|
DUOX2_HUMAN
|
||||||
| θ value | 4.28545e-26 (rank : 9) | NC score | 0.505885 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRD8, Q9NR02, Q9UHF9 | Gene names | DUOX2, LNOX2, THOX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 2 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH oxidase/peroxidase DUOX2) (NADPH thyroid oxidase 2) (Thyroid oxidase 2) (NADH/NADPH thyroid oxidase p138-tox) (p138 thyroid oxidase) (Large NOX 2) (Long NOX 2). | |||||
|
PGH1_MOUSE
|
||||||
| θ value | 1.69304e-06 (rank : 10) | NC score | 0.308215 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P22437 | Gene names | Ptgs1, Cox-1, Cox1 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin G/H synthase 1 precursor (EC 1.14.99.1) (Cyclooxygenase- 1) (COX-1) (Prostaglandin-endoperoxide synthase 1) (Prostaglandin H2 synthase 1) (PGH synthase 1) (PGHS-1) (PHS 1). | |||||
|
PGH1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 11) | NC score | 0.301236 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P23219, Q15122 | Gene names | PTGS1, COX1 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin G/H synthase 1 precursor (EC 1.14.99.1) (Cyclooxygenase- 1) (COX-1) (Prostaglandin-endoperoxide synthase 1) (Prostaglandin H2 synthase 1) (PGH synthase 1) (PGHS-1) (PHS 1). | |||||
|
PGH2_MOUSE
|
||||||
| θ value | 4.1701e-05 (rank : 12) | NC score | 0.280586 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q05769 | Gene names | Ptgs2, Cox-2, Cox2, Pghs-b, Tis10 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin G/H synthase 2 precursor (EC 1.14.99.1) (Cyclooxygenase- 2) (COX-2) (Prostaglandin-endoperoxide synthase 2) (Prostaglandin H2 synthase 2) (PGH synthase 2) (PGHS-2) (PHS II) (Glucocorticoid- regulated inflammatory cyclooxygenase) (Gripghs) (TIS10 protein) (Macrophage activation-associated marker protein P71/73) (PES-2). | |||||
|
PGH2_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 13) | NC score | 0.293475 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35354, Q16876 | Gene names | PTGS2, COX2 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin G/H synthase 2 precursor (EC 1.14.99.1) (Cyclooxygenase- 2) (COX-2) (Prostaglandin-endoperoxide synthase 2) (Prostaglandin H2 synthase 2) (PGH synthase 2) (PGHS-2) (PHS II). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 14) | NC score | 0.008987 (rank : 25) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
PPAL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.018623 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24638, Q8QZT5 | Gene names | Acp2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal acid phosphatase precursor (EC 3.1.3.2) (LAP). | |||||
|
MCM4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 16) | NC score | 0.008913 (rank : 26) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P33991, Q8NEH1, Q99658 | Gene names | MCM4, CDC21 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
TERF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 17) | NC score | 0.016402 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54274, Q15553, Q93029 | Gene names | TERF1, PIN2, TRF, TRF1 | |||
|
Domain Architecture |
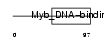
|
|||||
| Description | Telomeric repeat-binding factor 1 (TTAGGG repeat-binding factor 1) (NIMA-interacting protein 2) (Telomeric protein Pin2/TRF1). | |||||
|
ITSN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.006028 (rank : 29) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
DHDDS_MOUSE
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.014394 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KU1, Q3UF13, Q8BZ16, Q8BZK8 | Gene names | Dhdds | |||
|
Domain Architecture |
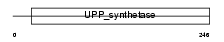
|
|||||
| Description | Dehydrodolichyl diphosphate synthase (EC 2.5.1.-) (Dedol-PP synthase). | |||||
|
DLGP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.007242 (rank : 28) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BJ42, Q6XBF3 | Gene names | Dlgap2, Dap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 2 (DAP-2) (SAP90/PSD-95-associated protein 2) (SAPAP2) (PSD-95/SAP90-binding protein 2). | |||||
|
OL490_MOUSE
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | -0.001630 (rank : 32) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 691 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VFD2 | Gene names | Olfr490, Mor204-17 | |||
|
Domain Architecture |
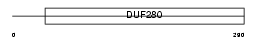
|
|||||
| Description | Olfactory receptor 490 (Olfactory receptor 204-17). | |||||
|
IGF1R_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.001297 (rank : 31) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08069 | Gene names | IGF1R | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) [Contains: Insulin-like growth factor 1 receptor alpha chain; Insulin-like growth factor 1 receptor beta chain]. | |||||
|
IGF1R_MOUSE
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.001305 (rank : 30) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60751, O70438, Q62123 | Gene names | Igf1r | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) [Contains: Insulin-like growth factor 1 receptor alpha chain; Insulin-like growth factor 1 receptor beta chain]. | |||||
|
K1683_HUMAN
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.008319 (rank : 27) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
CY24B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.069510 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04839, Q2PP16 | Gene names | CYBB, NOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome b-245 heavy chain (p22 phagocyte B-cytochrome) (Neutrophil cytochrome b 91 kDa polypeptide) (CGD91-phox) (gp91-phox) (gp91-1) (Heme-binding membrane glycoprotein gp91phox) (Cytochrome b(558) subunit beta) (Cytochrome b558 subunit beta) (Superoxide-generating NADPH oxidase heavy chain subunit) (NADPH oxidase 2). | |||||
|
CY24B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.069437 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61093 | Gene names | Cybb, Cgd | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome b-245 heavy chain (p22 phagocyte B-cytochrome) (Neutrophil cytochrome b 91 kDa polypeptide) (CGD91-phox) (gp91-phox) (gp91-1) (Heme-binding membrane glycoprotein gp91phox) (Cytochrome b(558) subunit beta) (Cytochrome b558 subunit beta). | |||||
|
NOX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.073966 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5S8, O95691, Q2PP02 | Gene names | NOX1, NOH1 | |||
|
Domain Architecture |
|
|||||
| Description | NADPH oxidase homolog 1 (NOX-1) (NOH-1) (NADH/NADPH mitogenic oxidase subunit P65-MOX) (Mitogenic oxidase 1) (MOX1). | |||||
|
NOX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.075397 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBY0, Q9HBJ9 | Gene names | NOX3, MOX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase 3 (EC 1.6.3.-) (gp91phox homolog 3) (GP91-3) (Mitogenic oxidase 2). | |||||
|
NOX3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.073711 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q672J9, Q6Y4Q8 | Gene names | Nox3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase 3 (EC 1.6.3.-). | |||||
|
NOX4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.064759 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPH5, Q5K3R4, Q5K3R5, Q5K3R6, Q5K3R8, Q7Z7G3, Q86V92 | Gene names | NOX4, RENOX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase 4 (EC 1.6.3.-) (Kidney superoxide-producing NADPH oxidase) (KOX-1) (Renal NAD(P)H-oxidase). | |||||
|
NOX4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.066152 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHI8, Q3TF39, Q8C3M1, Q8VCA3 | Gene names | Nox4, Renox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase 4 (EC 1.6.3.-) (Kidney superoxide-producing NADPH oxidase) (Kox-1) (Renal NAD(P)H-oxidase) (Superoxide-generating NADPH oxidase 4). | |||||
|
NOX5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.086606 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96PH1, Q8TEQ1, Q8TER4, Q96PH2, Q96PJ8, Q96PJ9, Q9H6E0, Q9HAM8 | Gene names | NOX5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase 5 (EC 1.6.3.-). | |||||
|
PERL_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P22079, Q13408 | Gene names | LPO, SAPX | |||
|
Domain Architecture |

|
|||||
| Description | Lactoperoxidase precursor (EC 1.11.1.7) (LPO) (Salivary peroxidase) (SPO). | |||||
|
PERM_MOUSE
|
||||||
| NC score | 0.992369 (rank : 2) | θ value | 0 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11247 | Gene names | Mpo | |||
|
Domain Architecture |

|
|||||
| Description | Myeloperoxidase precursor (EC 1.11.1.7) (MPO) [Contains: Myeloperoxidase light chain; Myeloperoxidase heavy chain]. | |||||
|
PERM_HUMAN
|
||||||
| NC score | 0.991810 (rank : 3) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P05164, Q14862, Q4PJH5 | Gene names | MPO | |||
|
Domain Architecture |

|
|||||
| Description | Myeloperoxidase precursor (EC 1.11.1.7) (MPO) [Contains: 89 kDa myeloperoxidase; 84 kDa myeloperoxidase; Myeloperoxidase light chain; Myeloperoxidase heavy chain]. | |||||
|
PERE_HUMAN
|
||||||
| NC score | 0.990515 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P11678, Q4TVP3 | Gene names | EPX, EPER, EPO, EPP | |||
|
Domain Architecture |

|
|||||
| Description | Eosinophil peroxidase precursor (EC 1.11.1.7) (EPO) [Contains: Eosinophil peroxidase light chain; Eosinophil peroxidase heavy chain]. | |||||
|
PERE_MOUSE
|
||||||
| NC score | 0.989455 (rank : 5) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P49290, Q61798 | Gene names | Epx, Eper | |||
|
Domain Architecture |

|
|||||
| Description | Eosinophil peroxidase precursor (EC 1.11.1.7) (EPO) [Contains: Eosinophil peroxidase light chain; Eosinophil peroxidase heavy chain]. | |||||
|
PERT_MOUSE
|
||||||
| NC score | 0.859008 (rank : 6) | θ value | 2.12077e-150 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35419, Q8C8B1 | Gene names | Tpo | |||
|
Domain Architecture |
|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
PERT_HUMAN
|
||||||
| NC score | 0.851853 (rank : 7) | θ value | 1.91815e-151 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P07202, P09934, P09935, Q8IUL0, Q8NF94, Q8NF95, Q8NF96, Q8NF97, Q8TCI9 | Gene names | TPO | |||
|
Domain Architecture |

|
|||||
| Description | Thyroid peroxidase precursor (EC 1.11.1.8) (TPO). | |||||
|
DUOX1_HUMAN
|
||||||
| NC score | 0.514553 (rank : 8) | θ value | 1.73987e-27 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRD9, Q6ZMB3, Q6ZR09, Q9NZC1 | Gene names | DUOX1, DUOX, LNOX1, THOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 1 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH thyroid oxidase 1) (Thyroid oxidase 1) (Large NOX 1) (Long NOX 1). | |||||
|
DUOX2_HUMAN
|
||||||
| NC score | 0.505885 (rank : 9) | θ value | 4.28545e-26 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NRD8, Q9NR02, Q9UHF9 | Gene names | DUOX2, LNOX2, THOX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 2 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH oxidase/peroxidase DUOX2) (NADPH thyroid oxidase 2) (Thyroid oxidase 2) (NADH/NADPH thyroid oxidase p138-tox) (p138 thyroid oxidase) (Large NOX 2) (Long NOX 2). | |||||
|
PGH1_MOUSE
|
||||||
| NC score | 0.308215 (rank : 10) | θ value | 1.69304e-06 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 100 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P22437 | Gene names | Ptgs1, Cox-1, Cox1 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin G/H synthase 1 precursor (EC 1.14.99.1) (Cyclooxygenase- 1) (COX-1) (Prostaglandin-endoperoxide synthase 1) (Prostaglandin H2 synthase 1) (PGH synthase 1) (PGHS-1) (PHS 1). | |||||
|
PGH1_HUMAN
|
||||||
| NC score | 0.301236 (rank : 11) | θ value | 8.40245e-06 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P23219, Q15122 | Gene names | PTGS1, COX1 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin G/H synthase 1 precursor (EC 1.14.99.1) (Cyclooxygenase- 1) (COX-1) (Prostaglandin-endoperoxide synthase 1) (Prostaglandin H2 synthase 1) (PGH synthase 1) (PGHS-1) (PHS 1). | |||||
|
PGH2_HUMAN
|
||||||
| NC score | 0.293475 (rank : 12) | θ value | 7.1131e-05 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 103 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P35354, Q16876 | Gene names | PTGS2, COX2 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin G/H synthase 2 precursor (EC 1.14.99.1) (Cyclooxygenase- 2) (COX-2) (Prostaglandin-endoperoxide synthase 2) (Prostaglandin H2 synthase 2) (PGH synthase 2) (PGHS-2) (PHS II). | |||||
|
PGH2_MOUSE
|
||||||
| NC score | 0.280586 (rank : 13) | θ value | 4.1701e-05 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q05769 | Gene names | Ptgs2, Cox-2, Cox2, Pghs-b, Tis10 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin G/H synthase 2 precursor (EC 1.14.99.1) (Cyclooxygenase- 2) (COX-2) (Prostaglandin-endoperoxide synthase 2) (Prostaglandin H2 synthase 2) (PGH synthase 2) (PGHS-2) (PHS II) (Glucocorticoid- regulated inflammatory cyclooxygenase) (Gripghs) (TIS10 protein) (Macrophage activation-associated marker protein P71/73) (PES-2). | |||||
|
NOX5_HUMAN
|
||||||
| NC score | 0.086606 (rank : 14) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96PH1, Q8TEQ1, Q8TER4, Q96PH2, Q96PJ8, Q96PJ9, Q9H6E0, Q9HAM8 | Gene names | NOX5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase 5 (EC 1.6.3.-). | |||||
|
NOX3_HUMAN
|
||||||
| NC score | 0.075397 (rank : 15) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HBY0, Q9HBJ9 | Gene names | NOX3, MOX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase 3 (EC 1.6.3.-) (gp91phox homolog 3) (GP91-3) (Mitogenic oxidase 2). | |||||
|
NOX1_HUMAN
|
||||||
| NC score | 0.073966 (rank : 16) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y5S8, O95691, Q2PP02 | Gene names | NOX1, NOH1 | |||
|
Domain Architecture |
|
|||||
| Description | NADPH oxidase homolog 1 (NOX-1) (NOH-1) (NADH/NADPH mitogenic oxidase subunit P65-MOX) (Mitogenic oxidase 1) (MOX1). | |||||
|
NOX3_MOUSE
|
||||||
| NC score | 0.073711 (rank : 17) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q672J9, Q6Y4Q8 | Gene names | Nox3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase 3 (EC 1.6.3.-). | |||||
|
CY24B_HUMAN
|
||||||
| NC score | 0.069510 (rank : 18) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04839, Q2PP16 | Gene names | CYBB, NOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome b-245 heavy chain (p22 phagocyte B-cytochrome) (Neutrophil cytochrome b 91 kDa polypeptide) (CGD91-phox) (gp91-phox) (gp91-1) (Heme-binding membrane glycoprotein gp91phox) (Cytochrome b(558) subunit beta) (Cytochrome b558 subunit beta) (Superoxide-generating NADPH oxidase heavy chain subunit) (NADPH oxidase 2). | |||||
|
CY24B_MOUSE
|
||||||
| NC score | 0.069437 (rank : 19) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61093 | Gene names | Cybb, Cgd | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome b-245 heavy chain (p22 phagocyte B-cytochrome) (Neutrophil cytochrome b 91 kDa polypeptide) (CGD91-phox) (gp91-phox) (gp91-1) (Heme-binding membrane glycoprotein gp91phox) (Cytochrome b(558) subunit beta) (Cytochrome b558 subunit beta). | |||||
|
NOX4_MOUSE
|
||||||
| NC score | 0.066152 (rank : 20) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JHI8, Q3TF39, Q8C3M1, Q8VCA3 | Gene names | Nox4, Renox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase 4 (EC 1.6.3.-) (Kidney superoxide-producing NADPH oxidase) (Kox-1) (Renal NAD(P)H-oxidase) (Superoxide-generating NADPH oxidase 4). | |||||
|
NOX4_HUMAN
|
||||||
| NC score | 0.064759 (rank : 21) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NPH5, Q5K3R4, Q5K3R5, Q5K3R6, Q5K3R8, Q7Z7G3, Q86V92 | Gene names | NOX4, RENOX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase 4 (EC 1.6.3.-) (Kidney superoxide-producing NADPH oxidase) (KOX-1) (Renal NAD(P)H-oxidase). | |||||
|
PPAL_MOUSE
|
||||||
| NC score | 0.018623 (rank : 22) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P24638, Q8QZT5 | Gene names | Acp2 | |||
|
Domain Architecture |

|
|||||
| Description | Lysosomal acid phosphatase precursor (EC 3.1.3.2) (LAP). | |||||
|
TERF1_HUMAN
|
||||||
| NC score | 0.016402 (rank : 23) | θ value | 3.0926 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P54274, Q15553, Q93029 | Gene names | TERF1, PIN2, TRF, TRF1 | |||
|
Domain Architecture |

|
|||||
| Description | Telomeric repeat-binding factor 1 (TTAGGG repeat-binding factor 1) (NIMA-interacting protein 2) (Telomeric protein Pin2/TRF1). | |||||
|
DHDDS_MOUSE
|
||||||
| NC score | 0.014394 (rank : 24) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KU1, Q3UF13, Q8BZ16, Q8BZK8 | Gene names | Dhdds | |||
|
Domain Architecture |

|
|||||
| Description | Dehydrodolichyl diphosphate synthase (EC 2.5.1.-) (Dedol-PP synthase). | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.008987 (rank : 25) | θ value | 0.21417 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
MCM4_HUMAN
|
||||||
| NC score | 0.008913 (rank : 26) | θ value | 3.0926 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P33991, Q8NEH1, Q99658 | Gene names | MCM4, CDC21 | |||
|
Domain Architecture |

|
|||||
| Description | DNA replication licensing factor MCM4 (CDC21 homolog) (P1-CDC21). | |||||
|
K1683_HUMAN
|
||||||
| NC score | 0.008319 (rank : 27) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 240 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0B3, Q8N4G8, Q96M14, Q9C0I0 | Gene names | KIAA1683 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1683. | |||||
|
DLGP2_MOUSE
|
||||||
| NC score | 0.007242 (rank : 28) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BJ42, Q6XBF3 | Gene names | Dlgap2, Dap2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Disks large-associated protein 2 (DAP-2) (SAP90/PSD-95-associated protein 2) (SAPAP2) (PSD-95/SAP90-binding protein 2). | |||||
|
ITSN1_HUMAN
|
||||||
| NC score | 0.006028 (rank : 29) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1757 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15811, O95216, Q9UET5, Q9UK60, Q9UNK1, Q9UNK2, Q9UQ92 | Gene names | ITSN1, ITSN, SH3D1A | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (SH3 domain-containing protein 1A) (SH3P17). | |||||
|
IGF1R_MOUSE
|
||||||
| NC score | 0.001305 (rank : 30) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60751, O70438, Q62123 | Gene names | Igf1r | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) [Contains: Insulin-like growth factor 1 receptor alpha chain; Insulin-like growth factor 1 receptor beta chain]. | |||||
|
IGF1R_HUMAN
|
||||||
| NC score | 0.001297 (rank : 31) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08069 | Gene names | IGF1R | |||
|
Domain Architecture |

|
|||||
| Description | Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) [Contains: Insulin-like growth factor 1 receptor alpha chain; Insulin-like growth factor 1 receptor beta chain]. | |||||
|
OL490_MOUSE
|
||||||
| NC score | -0.001630 (rank : 32) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 691 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VFD2 | Gene names | Olfr490, Mor204-17 | |||
|
Domain Architecture |

|
|||||
| Description | Olfactory receptor 490 (Olfactory receptor 204-17). | |||||