Please be patient as the page loads
|
P3_MOUSE
|
||||||
| SwissProt Accessions | P21129, Q8QZR2 | Gene names | Slc10a3, P3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P3 protein (Solute carrier family 10 member 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
P3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.997570 (rank : 2) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P09131 | Gene names | SLC10A3, DXS253E, P3 | |||
|
Domain Architecture |
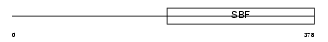
|
|||||
| Description | P3 protein (Solute carrier family 10 member 3). | |||||
|
P3_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P21129, Q8QZR2 | Gene names | Slc10a3, P3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P3 protein (Solute carrier family 10 member 3). | |||||
|
NTCP_HUMAN
|
||||||
| θ value | 1.2049e-28 (rank : 3) | NC score | 0.793666 (rank : 3) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14973 | Gene names | SLC10A1, NTCP | |||
|
Domain Architecture |
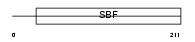
|
|||||
| Description | Sodium/bile acid cotransporter (Na(+)/bile acid cotransporter) (Na(+)/taurocholate transport protein) (Sodium/taurocholate cotransporting polypeptide) (Solute carrier family 10 member 1). | |||||
|
NTCP_MOUSE
|
||||||
| θ value | 3.28125e-26 (rank : 4) | NC score | 0.788222 (rank : 4) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08705, Q52L86 | Gene names | Slc10a1, Ntcp | |||
|
Domain Architecture |
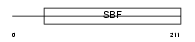
|
|||||
| Description | Sodium/bile acid cotransporter (Na(+)/bile acid cotransporter) (Na(+)/taurocholate transport protein) (Sodium/taurocholate cotransporting polypeptide) (Solute carrier family 10 member 1). | |||||
|
NTCP2_MOUSE
|
||||||
| θ value | 1.42661e-21 (rank : 5) | NC score | 0.749074 (rank : 5) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70172, Q8VI83, Q925U7 | Gene names | Slc10a2, Ntcp2 | |||
|
Domain Architecture |
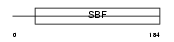
|
|||||
| Description | Ileal sodium/bile acid cotransporter (Ileal Na(+)/bile acid cotransporter) (Na(+)-dependent ileal bile acid transporter) (Ileal sodium-dependent bile acid transporter) (ISBT) (IBAT) (Apical sodium- dependent bile acid transporter) (ASBT) (Sodium/taurocholate cotransporting polypeptide, ileal) (Solute carrier family 10 member 2). | |||||
|
NTCP2_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 6) | NC score | 0.737797 (rank : 6) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12908, Q13839 | Gene names | SLC10A2, NTCP2 | |||
|
Domain Architecture |
|
|||||
| Description | Ileal sodium/bile acid cotransporter (Ileal Na(+)/bile acid cotransporter) (Na(+)-dependent ileal bile acid transporter) (Ileal sodium-dependent bile acid transporter) (ISBT) (IBAT) (Apical sodium- dependent bile acid transporter) (ASBT) (Sodium/taurocholate cotransporting polypeptide, ileal) (Solute carrier family 10 member 2). | |||||
|
G137B_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 7) | NC score | 0.137897 (rank : 7) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60478 | Gene names | GPR137B, TM7SF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR137B (Transmembrane 7 superfamily member 1). | |||||
|
CLD2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 8) | NC score | 0.020583 (rank : 8) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57739 | Gene names | CLDN2 | |||
|
Domain Architecture |
|
|||||
| Description | Claudin-2 (SP82). | |||||
|
CLD2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 9) | NC score | 0.018919 (rank : 9) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88552 | Gene names | Cldn2 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-2. | |||||
|
CCRK_HUMAN
|
||||||
| θ value | 8.99809 (rank : 10) | NC score | -0.000144 (rank : 10) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IZL9, Q5EDC4, Q5VYW1, Q9BUF4 | Gene names | CCRK, CDCH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle-related kinase (EC 2.7.11.22) (Cyclin-kinase-activating kinase p42) (CDK-activating kinase p42) (CAK-kinase p42) (Cyclin- dependent protein kinase H). | |||||
|
P3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P21129, Q8QZR2 | Gene names | Slc10a3, P3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | P3 protein (Solute carrier family 10 member 3). | |||||
|
P3_HUMAN
|
||||||
| NC score | 0.997570 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P09131 | Gene names | SLC10A3, DXS253E, P3 | |||
|
Domain Architecture |

|
|||||
| Description | P3 protein (Solute carrier family 10 member 3). | |||||
|
NTCP_HUMAN
|
||||||
| NC score | 0.793666 (rank : 3) | θ value | 1.2049e-28 (rank : 3) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q14973 | Gene names | SLC10A1, NTCP | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/bile acid cotransporter (Na(+)/bile acid cotransporter) (Na(+)/taurocholate transport protein) (Sodium/taurocholate cotransporting polypeptide) (Solute carrier family 10 member 1). | |||||
|
NTCP_MOUSE
|
||||||
| NC score | 0.788222 (rank : 4) | θ value | 3.28125e-26 (rank : 4) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O08705, Q52L86 | Gene names | Slc10a1, Ntcp | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/bile acid cotransporter (Na(+)/bile acid cotransporter) (Na(+)/taurocholate transport protein) (Sodium/taurocholate cotransporting polypeptide) (Solute carrier family 10 member 1). | |||||
|
NTCP2_MOUSE
|
||||||
| NC score | 0.749074 (rank : 5) | θ value | 1.42661e-21 (rank : 5) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P70172, Q8VI83, Q925U7 | Gene names | Slc10a2, Ntcp2 | |||
|
Domain Architecture |

|
|||||
| Description | Ileal sodium/bile acid cotransporter (Ileal Na(+)/bile acid cotransporter) (Na(+)-dependent ileal bile acid transporter) (Ileal sodium-dependent bile acid transporter) (ISBT) (IBAT) (Apical sodium- dependent bile acid transporter) (ASBT) (Sodium/taurocholate cotransporting polypeptide, ileal) (Solute carrier family 10 member 2). | |||||
|
NTCP2_HUMAN
|
||||||
| NC score | 0.737797 (rank : 6) | θ value | 3.28887e-18 (rank : 6) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q12908, Q13839 | Gene names | SLC10A2, NTCP2 | |||
|
Domain Architecture |

|
|||||
| Description | Ileal sodium/bile acid cotransporter (Ileal Na(+)/bile acid cotransporter) (Na(+)-dependent ileal bile acid transporter) (Ileal sodium-dependent bile acid transporter) (ISBT) (IBAT) (Apical sodium- dependent bile acid transporter) (ASBT) (Sodium/taurocholate cotransporting polypeptide, ileal) (Solute carrier family 10 member 2). | |||||
|
G137B_HUMAN
|
||||||
| NC score | 0.137897 (rank : 7) | θ value | 0.0563607 (rank : 7) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60478 | Gene names | GPR137B, TM7SF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integral membrane protein GPR137B (Transmembrane 7 superfamily member 1). | |||||
|
CLD2_HUMAN
|
||||||
| NC score | 0.020583 (rank : 8) | θ value | 1.81305 (rank : 8) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57739 | Gene names | CLDN2 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-2 (SP82). | |||||
|
CLD2_MOUSE
|
||||||
| NC score | 0.018919 (rank : 9) | θ value | 3.0926 (rank : 9) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88552 | Gene names | Cldn2 | |||
|
Domain Architecture |

|
|||||
| Description | Claudin-2. | |||||
|
CCRK_HUMAN
|
||||||
| NC score | -0.000144 (rank : 10) | θ value | 8.99809 (rank : 10) | |||
| Query Neighborhood Hits | 10 | Target Neighborhood Hits | 835 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IZL9, Q5EDC4, Q5VYW1, Q9BUF4 | Gene names | CCRK, CDCH | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell cycle-related kinase (EC 2.7.11.22) (Cyclin-kinase-activating kinase p42) (CDK-activating kinase p42) (CAK-kinase p42) (Cyclin- dependent protein kinase H). | |||||