Please be patient as the page loads
|
OAS3_HUMAN
|
||||||
| SwissProt Accessions | Q9Y6K5, Q9H3P5 | Gene names | OAS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 3 (EC 2.7.7.-) ((2-5')oligo(A) synthetase 3) (2-5A synthetase 3) (p100 OAS) (p100OAS). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
OAS3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y6K5, Q9H3P5 | Gene names | OAS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 3 (EC 2.7.7.-) ((2-5')oligo(A) synthetase 3) (2-5A synthetase 3) (p100 OAS) (p100OAS). | |||||
|
OAS2_HUMAN
|
||||||
| θ value | 2.18449e-163 (rank : 2) | NC score | 0.967095 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29728, Q86XX8 | Gene names | OAS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 2 (EC 2.7.7.-) ((2-5')oligo(A) synthetase 2) (2-5A synthetase 2) (p69 OAS / p71 OAS) (p69OAS / p71OAS). | |||||
|
OAS1_HUMAN
|
||||||
| θ value | 2.22025e-107 (rank : 3) | NC score | 0.957894 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P00973, P04820, P29080, P29081, P78485, P78486, Q16700, Q16701, Q96J61 | Gene names | OAS1, OIAS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 1 (EC 2.7.7.-) ((2-5')oligo(A) synthetase 1) (2-5A synthetase 1) (p46/p42 OAS) (E18/E16). | |||||
|
OAS1A_MOUSE
|
||||||
| θ value | 1.54472e-92 (rank : 4) | NC score | 0.953654 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11928, Q64440 | Gene names | Oas1a, Oias1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 1A (EC 2.7.7.-) ((2-5')oligo(A) synthetase 1A) (2-5A synthetase 1A) (p42 OAS). | |||||
|
OASL2_MOUSE
|
||||||
| θ value | 2.82823e-78 (rank : 5) | NC score | 0.928938 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2F2 | Gene names | Oasl2 | |||
|
Domain Architecture |

|
|||||
| Description | 54 kDa 2'-5'-oligoadenylate synthetase-like protein 2 (EC 2.7.7.-) (p54 OASL) (p54OASL) (M1204). | |||||
|
OASL_HUMAN
|
||||||
| θ value | 4.37348e-71 (rank : 6) | NC score | 0.901551 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15646, O75686, Q9Y6K6, Q9Y6K7 | Gene names | OASL, TRIP14 | |||
|
Domain Architecture |

|
|||||
| Description | 59 kDa 2'-5'-oligoadenylate synthetase-like protein (p59 OASL) (p59OASL) (Thyroid receptor-interacting protein 14) (TRIP14). | |||||
|
RBM12_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 7) | NC score | 0.054191 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
MYT1_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.041355 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
SMAL1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.030503 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
TB182_HUMAN
|
||||||
| θ value | 0.163984 (rank : 10) | NC score | 0.048786 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
EXTL3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.025737 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVL6 | Gene names | Extl3 | |||
|
Domain Architecture |

|
|||||
| Description | Exostosin-like 3 (EC 2.4.1.-) (Multiple exostosis-like protein 3). | |||||
|
HXD13_HUMAN
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.012165 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35453 | Gene names | HOXD13, HOX4I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D13 (Hox-4I). | |||||
|
PCQAP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 13) | NC score | 0.026231 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
MOD5_MOUSE
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.029138 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80UN9, Q9D1H5 | Gene names | Trit1, Ipt | |||
|
Domain Architecture |
|
|||||
| Description | tRNA isopentenyltransferase, mitochondrial precursor (EC 2.5.1.8) (Isopentenyl-diphosphate:tRNA isopentenyltransferase) (IPP transferase) (IPTase) (IPPT). | |||||
|
MYT1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.033044 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
PHLPP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.008414 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
UBP36_HUMAN
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.012073 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
DYN3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.010995 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
RX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.006186 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2V3 | Gene names | RAX, RX | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
CKLF2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.024872 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAZ6, Q8N7E5 | Gene names | CMTM2, CKLFSF2 | |||
|
Domain Architecture |
|
|||||
| Description | CKLF-like MARVEL transmembrane domain-containing protein 2 (Chemokine- like factor superfamily member 2). | |||||
|
HD_MOUSE
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.016435 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
LTBP3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.004175 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61810, Q8BNQ6 | Gene names | Ltbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
VMDL1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.012292 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGM5, Q8VCM0 | Gene names | Vmd2l1 | |||
|
Domain Architecture |

|
|||||
| Description | Bestrophin-2 (Vitelliform macular dystrophy 2-like protein 1). | |||||
|
ANX11_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.006797 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
BRCA2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 25) | NC score | 0.014126 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
CN004_MOUSE
|
||||||
| θ value | 5.27518 (rank : 26) | NC score | 0.017279 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
DDX11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 27) | NC score | 0.013015 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96FC9, Q13333, Q86VQ4, Q86W62, Q92498, Q92770, Q92998, Q92999 | Gene names | DDX11, CHL1, CHLR1, KRG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX11 (EC 3.6.1.-) (DEAD/H box protein 11) (CHL1 homolog) (Keratinocyte growth factor-regulated gene 2 protein) (KRG-2). | |||||
|
MORC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.010805 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6X9, Q9UF28, Q9Y6V2 | Gene names | MORC2, KIAA0852, ZCWCC1 | |||
|
Domain Architecture |

|
|||||
| Description | MORC family CW-type zinc finger protein 2 (Zinc finger CW-type coiled- coil domain protein 1). | |||||
|
SRBS1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.009638 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
CT055_HUMAN
|
||||||
| θ value | 6.88961 (rank : 30) | NC score | 0.017929 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQ89 | Gene names | C20orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf55. | |||||
|
DOT1L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.014081 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
FOXGB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.004638 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55315 | Gene names | FOXG1B, FKHL1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1B (Forkhead-related protein FKHL1) (Transcription factor BF-1) (Brain factor 1) (BF1) (HFK1). | |||||
|
IGHG4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.004779 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01861 | Gene names | IGHG4 | |||
|
Domain Architecture |
|
|||||
| Description | Ig gamma-4 chain C region. | |||||
|
MYT1L_HUMAN
|
||||||
| θ value | 6.88961 (rank : 34) | NC score | 0.025607 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
PCLO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.008784 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PCLO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.009502 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PMS1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.027918 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
TBX3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.005799 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15119, Q8TB20, Q9UKF8 | Gene names | TBX3 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX3 (T-box protein 3). | |||||
|
ADCY1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 39) | NC score | 0.004463 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08828, Q75MI1 | Gene names | ADCY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
CCKN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 40) | NC score | 0.018001 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09240 | Gene names | Cck | |||
|
Domain Architecture |
|
|||||
| Description | Cholecystokinins precursor (CCK) [Contains: Cholecystokinin 33 (CCK33); Cholecystokinin 12 (CCK12); Cholecystokinin 8 (CCK8)]. | |||||
|
DBND2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 41) | NC score | 0.015037 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQY9, Q9BQZ0, Q9BVL1, Q9H1F6, Q9NWZ0, Q9NY07, Q9NZ31 | Gene names | DBNDD2, C20orf35 | |||
|
Domain Architecture |
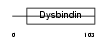
|
|||||
| Description | Dysbindin domain-containing protein 2 (HSMNP1). | |||||
|
EGR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 42) | NC score | -0.000769 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08152 | Gene names | Egr2, Egr-2, Krox-20, Zfp-25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein). | |||||
|
EHMT2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 43) | NC score | 0.002072 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
EXTL3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 44) | NC score | 0.018639 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43909, O00225 | Gene names | EXTL3, EXTL1L, EXTR1, KIAA0519 | |||
|
Domain Architecture |

|
|||||
| Description | Exostosin-like 3 (EC 2.4.1.223) (Glucuronyl-galactosyl-proteoglycan 4- alpha-N-acetylglucosaminyltransferase) (Putative tumor suppressor protein EXTL3) (Multiple exostosis-like protein 3) (Hereditary multiple exostoses gene isolog) (EXT-related protein 1). | |||||
|
F113A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 45) | NC score | 0.012615 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H1Q7, Q5JUA5, Q5JUA6, Q6PK19, Q86WF5, Q96CG7, Q9H1Q6, Q9H6D1 | Gene names | FAM113A, C20orf81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM113A (Sarcoma antigen NY-SAR-23). | |||||
|
MTP_MOUSE
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.011290 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08601 | Gene names | Mttp, Mtp | |||
|
Domain Architecture |
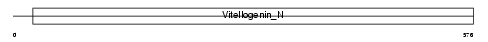
|
|||||
| Description | Microsomal triglyceride transfer protein large subunit precursor. | |||||
|
SSBP4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.009686 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
WNK1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.000406 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.000841 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
OAS3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9Y6K5, Q9H3P5 | Gene names | OAS3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 3 (EC 2.7.7.-) ((2-5')oligo(A) synthetase 3) (2-5A synthetase 3) (p100 OAS) (p100OAS). | |||||
|
OAS2_HUMAN
|
||||||
| NC score | 0.967095 (rank : 2) | θ value | 2.18449e-163 (rank : 2) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P29728, Q86XX8 | Gene names | OAS2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 2 (EC 2.7.7.-) ((2-5')oligo(A) synthetase 2) (2-5A synthetase 2) (p69 OAS / p71 OAS) (p69OAS / p71OAS). | |||||
|
OAS1_HUMAN
|
||||||
| NC score | 0.957894 (rank : 3) | θ value | 2.22025e-107 (rank : 3) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P00973, P04820, P29080, P29081, P78485, P78486, Q16700, Q16701, Q96J61 | Gene names | OAS1, OIAS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 1 (EC 2.7.7.-) ((2-5')oligo(A) synthetase 1) (2-5A synthetase 1) (p46/p42 OAS) (E18/E16). | |||||
|
OAS1A_MOUSE
|
||||||
| NC score | 0.953654 (rank : 4) | θ value | 1.54472e-92 (rank : 4) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P11928, Q64440 | Gene names | Oas1a, Oias1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 2'-5'-oligoadenylate synthetase 1A (EC 2.7.7.-) ((2-5')oligo(A) synthetase 1A) (2-5A synthetase 1A) (p42 OAS). | |||||
|
OASL2_MOUSE
|
||||||
| NC score | 0.928938 (rank : 5) | θ value | 2.82823e-78 (rank : 5) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z2F2 | Gene names | Oasl2 | |||
|
Domain Architecture |

|
|||||
| Description | 54 kDa 2'-5'-oligoadenylate synthetase-like protein 2 (EC 2.7.7.-) (p54 OASL) (p54OASL) (M1204). | |||||
|
OASL_HUMAN
|
||||||
| NC score | 0.901551 (rank : 6) | θ value | 4.37348e-71 (rank : 6) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q15646, O75686, Q9Y6K6, Q9Y6K7 | Gene names | OASL, TRIP14 | |||
|
Domain Architecture |

|
|||||
| Description | 59 kDa 2'-5'-oligoadenylate synthetase-like protein (p59 OASL) (p59OASL) (Thyroid receptor-interacting protein 14) (TRIP14). | |||||
|
RBM12_MOUSE
|
||||||
| NC score | 0.054191 (rank : 7) | θ value | 0.00869519 (rank : 7) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
TB182_HUMAN
|
||||||
| NC score | 0.048786 (rank : 8) | θ value | 0.163984 (rank : 10) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
MYT1_HUMAN
|
||||||
| NC score | 0.041355 (rank : 9) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q01538, O94922, Q9UPV2 | Gene names | MYT1, KIAA0835, KIAA1050, MTF1, MYTI, PLPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Myelin transcription factor 1 (MyT1) (MyTI) (Proteolipid protein- binding protein) (PLPB1). | |||||
|
MYT1_MOUSE
|
||||||
| NC score | 0.033044 (rank : 10) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CFC2, O08995, Q8CFH1 | Gene names | Myt1, Kiaa0835, Nzf2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1 (MyT1) (Neural zinc finger factor 2) (NZF-2). | |||||
|
SMAL1_HUMAN
|
||||||
| NC score | 0.030503 (rank : 11) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9NZC9, Q96AY1, Q9NXQ5, Q9UFH3, Q9UI93 | Gene names | SMARCAL1, HARP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 (EC 3.6.1.-) (Sucrose nonfermenting protein 2-like 1) (HepA-related protein) (hHARP). | |||||
|
MOD5_MOUSE
|
||||||
| NC score | 0.029138 (rank : 12) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q80UN9, Q9D1H5 | Gene names | Trit1, Ipt | |||
|
Domain Architecture |

|
|||||
| Description | tRNA isopentenyltransferase, mitochondrial precursor (EC 2.5.1.8) (Isopentenyl-diphosphate:tRNA isopentenyltransferase) (IPP transferase) (IPTase) (IPPT). | |||||
|
PMS1_HUMAN
|
||||||
| NC score | 0.027918 (rank : 13) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P54277 | Gene names | PMS1, PMSL1 | |||
|
Domain Architecture |

|
|||||
| Description | PMS1 protein homolog 1 (DNA mismatch repair protein PMS1). | |||||
|
PCQAP_MOUSE
|
||||||
| NC score | 0.026231 (rank : 14) | θ value | 1.06291 (rank : 13) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q924H2 | Gene names | Pcqap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Positive cofactor 2 glutamine/Q-rich-associated protein (PC2 glutamine/Q-rich-associated protein) (mPcqap). | |||||
|
EXTL3_MOUSE
|
||||||
| NC score | 0.025737 (rank : 15) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVL6 | Gene names | Extl3 | |||
|
Domain Architecture |

|
|||||
| Description | Exostosin-like 3 (EC 2.4.1.-) (Multiple exostosis-like protein 3). | |||||
|
MYT1L_HUMAN
|
||||||
| NC score | 0.025607 (rank : 16) | θ value | 6.88961 (rank : 34) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UL68, Q6IQ17, Q9UPP6 | Gene names | MYT1L, KIAA1106 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myelin transcription factor 1-like protein (MyT1L protein) (MyT1-L). | |||||
|
CKLF2_HUMAN
|
||||||
| NC score | 0.024872 (rank : 17) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAZ6, Q8N7E5 | Gene names | CMTM2, CKLFSF2 | |||
|
Domain Architecture |

|
|||||
| Description | CKLF-like MARVEL transmembrane domain-containing protein 2 (Chemokine- like factor superfamily member 2). | |||||
|
EXTL3_HUMAN
|
||||||
| NC score | 0.018639 (rank : 18) | θ value | 8.99809 (rank : 44) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43909, O00225 | Gene names | EXTL3, EXTL1L, EXTR1, KIAA0519 | |||
|
Domain Architecture |

|
|||||
| Description | Exostosin-like 3 (EC 2.4.1.223) (Glucuronyl-galactosyl-proteoglycan 4- alpha-N-acetylglucosaminyltransferase) (Putative tumor suppressor protein EXTL3) (Multiple exostosis-like protein 3) (Hereditary multiple exostoses gene isolog) (EXT-related protein 1). | |||||
|
CCKN_MOUSE
|
||||||
| NC score | 0.018001 (rank : 19) | θ value | 8.99809 (rank : 40) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09240 | Gene names | Cck | |||
|
Domain Architecture |

|
|||||
| Description | Cholecystokinins precursor (CCK) [Contains: Cholecystokinin 33 (CCK33); Cholecystokinin 12 (CCK12); Cholecystokinin 8 (CCK8)]. | |||||
|
CT055_HUMAN
|
||||||
| NC score | 0.017929 (rank : 20) | θ value | 6.88961 (rank : 30) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQ89 | Gene names | C20orf55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C20orf55. | |||||
|
CN004_MOUSE
|
||||||
| NC score | 0.017279 (rank : 21) | θ value | 5.27518 (rank : 26) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
HD_MOUSE
|
||||||
| NC score | 0.016435 (rank : 22) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P42859 | Gene names | Hd, Hdh | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin (Huntington disease protein homolog) (HD protein). | |||||
|
DBND2_HUMAN
|
||||||
| NC score | 0.015037 (rank : 23) | θ value | 8.99809 (rank : 41) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BQY9, Q9BQZ0, Q9BVL1, Q9H1F6, Q9NWZ0, Q9NY07, Q9NZ31 | Gene names | DBNDD2, C20orf35 | |||
|
Domain Architecture |

|
|||||
| Description | Dysbindin domain-containing protein 2 (HSMNP1). | |||||
|
BRCA2_HUMAN
|
||||||
| NC score | 0.014126 (rank : 24) | θ value | 5.27518 (rank : 25) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P51587, O00183, O15008, Q13879 | Gene names | BRCA2, FANCD1 | |||
|
Domain Architecture |

|
|||||
| Description | Breast cancer type 2 susceptibility protein (Fanconi anemia group D1 protein). | |||||
|
DOT1L_HUMAN
|
||||||
| NC score | 0.014081 (rank : 25) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 660 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8TEK3, O60379, Q96JL1 | Gene names | DOT1L, KIAA1814 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-79 specific (EC 2.1.1.43) (Histone H3-K79 methyltransferase) (H3-K79-HMTase) (DOT1-like protein). | |||||
|
DDX11_HUMAN
|
||||||
| NC score | 0.013015 (rank : 26) | θ value | 5.27518 (rank : 27) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96FC9, Q13333, Q86VQ4, Q86W62, Q92498, Q92770, Q92998, Q92999 | Gene names | DDX11, CHL1, CHLR1, KRG2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ATP-dependent RNA helicase DDX11 (EC 3.6.1.-) (DEAD/H box protein 11) (CHL1 homolog) (Keratinocyte growth factor-regulated gene 2 protein) (KRG-2). | |||||
|
F113A_HUMAN
|
||||||
| NC score | 0.012615 (rank : 27) | θ value | 8.99809 (rank : 45) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H1Q7, Q5JUA5, Q5JUA6, Q6PK19, Q86WF5, Q96CG7, Q9H1Q6, Q9H6D1 | Gene names | FAM113A, C20orf81 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM113A (Sarcoma antigen NY-SAR-23). | |||||
|
VMDL1_MOUSE
|
||||||
| NC score | 0.012292 (rank : 28) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BGM5, Q8VCM0 | Gene names | Vmd2l1 | |||
|
Domain Architecture |

|
|||||
| Description | Bestrophin-2 (Vitelliform macular dystrophy 2-like protein 1). | |||||
|
HXD13_HUMAN
|
||||||
| NC score | 0.012165 (rank : 29) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 332 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35453 | Gene names | HOXD13, HOX4I | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-D13 (Hox-4I). | |||||
|
UBP36_HUMAN
|
||||||
| NC score | 0.012073 (rank : 30) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P275, Q8NDM8, Q9NVC8 | Gene names | USP36, KIAA1453 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 36 (EC 3.1.2.15) (Ubiquitin thioesterase 36) (Ubiquitin-specific-processing protease 36) (Deubiquitinating enzyme 36). | |||||
|
MTP_MOUSE
|
||||||
| NC score | 0.011290 (rank : 31) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08601 | Gene names | Mttp, Mtp | |||
|
Domain Architecture |
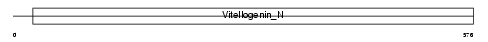
|
|||||
| Description | Microsomal triglyceride transfer protein large subunit precursor. | |||||
|
DYN3_HUMAN
|
||||||
| NC score | 0.010995 (rank : 32) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UQ16, O14982, O95555, Q5W129, Q9H0P3, Q9H548, Q9NQ68, Q9NQN6 | Gene names | DNM3, KIAA0820 | |||
|
Domain Architecture |

|
|||||
| Description | Dynamin-3 (EC 3.6.5.5) (Dynamin, testicular) (T-dynamin). | |||||
|
MORC2_HUMAN
|
||||||
| NC score | 0.010805 (rank : 33) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y6X9, Q9UF28, Q9Y6V2 | Gene names | MORC2, KIAA0852, ZCWCC1 | |||
|
Domain Architecture |

|
|||||
| Description | MORC family CW-type zinc finger protein 2 (Zinc finger CW-type coiled- coil domain protein 1). | |||||
|
SSBP4_HUMAN
|
||||||
| NC score | 0.009686 (rank : 34) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9BWG4 | Gene names | SSBP4 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 4. | |||||
|
SRBS1_HUMAN
|
||||||
| NC score | 0.009638 (rank : 35) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BX66, Q7LBE5, Q8IVK0, Q8IVQ4, Q96KF3, Q96KF4, Q9BX64, Q9BX65, Q9P2Q0, Q9UFT2, Q9UHN7, Q9Y338 | Gene names | SORBS1, KIAA1296, SH3D5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorbin and SH3 domain-containing protein 1 (Ponsin) (c-Cbl-associated protein) (CAP) (SH3 domain protein 5) (SH3P12). | |||||
|
PCLO_MOUSE
|
||||||
| NC score | 0.009502 (rank : 36) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2275 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q9QYX7, Q8CF91, Q8CF92, Q9QYX6, Q9QZJ0 | Gene names | Pclo, Acz | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin) (Multidomain presynaptic cytomatrix protein) (Brain-derived HLMN protein). | |||||
|
PCLO_HUMAN
|
||||||
| NC score | 0.008784 (rank : 37) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 2046 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9Y6V0, O43373, O60305, Q9BVC8, Q9UIV2, Q9Y6U9 | Gene names | PCLO, ACZ, KIAA0559 | |||
|
Domain Architecture |

|
|||||
| Description | Protein piccolo (Aczonin). | |||||
|
PHLPP_HUMAN
|
||||||
| NC score | 0.008414 (rank : 38) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 604 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O60346, Q641Q7, Q6P4C4, Q6PJI6, Q86TN6, Q96FK2, Q9NUY1 | Gene names | PHLPP, KIAA0606, PLEKHE1, SCOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PH domain leucine-rich repeat-containing protein phosphatase (EC 3.1.3.16) (PH domain leucine-rich repeat protein phosphatase) (Pleckstrin homology domain-containing family E protein 1) (Suprachiasmatic nucleus circadian oscillatory protein) (hSCOP). | |||||
|
ANX11_MOUSE
|
||||||
| NC score | 0.006797 (rank : 39) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
RX_HUMAN
|
||||||
| NC score | 0.006186 (rank : 40) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 423 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y2V3 | Gene names | RAX, RX | |||
|
Domain Architecture |

|
|||||
| Description | Retinal homeobox protein Rx (Retina and anterior neural fold homeobox protein). | |||||
|
TBX3_HUMAN
|
||||||
| NC score | 0.005799 (rank : 41) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15119, Q8TB20, Q9UKF8 | Gene names | TBX3 | |||
|
Domain Architecture |

|
|||||
| Description | T-box transcription factor TBX3 (T-box protein 3). | |||||
|
IGHG4_HUMAN
|
||||||
| NC score | 0.004779 (rank : 42) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P01861 | Gene names | IGHG4 | |||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-4 chain C region. | |||||
|
FOXGB_HUMAN
|
||||||
| NC score | 0.004638 (rank : 43) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55315 | Gene names | FOXG1B, FKHL1 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein G1B (Forkhead-related protein FKHL1) (Transcription factor BF-1) (Brain factor 1) (BF1) (HFK1). | |||||
|
ADCY1_HUMAN
|
||||||
| NC score | 0.004463 (rank : 44) | θ value | 8.99809 (rank : 39) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q08828, Q75MI1 | Gene names | ADCY1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Adenylate cyclase type 1 (EC 4.6.1.1) (Adenylate cyclase type I) (ATP pyrophosphate-lyase 1) (Ca(2+)/calmodulin-activated adenylyl cyclase). | |||||
|
LTBP3_MOUSE
|
||||||
| NC score | 0.004175 (rank : 45) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 475 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61810, Q8BNQ6 | Gene names | Ltbp3 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 3 precursor (LTBP-3). | |||||
|
EHMT2_HUMAN
|
||||||
| NC score | 0.002072 (rank : 46) | θ value | 8.99809 (rank : 43) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96KQ7, Q14349, Q6PK06, Q96MH5, Q96QD0, Q9UQL8, Q9Y331 | Gene names | EHMT2, BAT8, G9A, NG36 | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 3) (H3-K9-HMTase 3) (Euchromatic histone-lysine N-methyltransferase 2) (HLA-B-associated transcript 8) (Protein G9a). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.000841 (rank : 47) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
WNK1_HUMAN
|
||||||
| NC score | 0.000406 (rank : 48) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 1158 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H4A3, O15052, Q86WL5, Q8N673, Q9P1S9 | Gene names | WNK1, KDP, KIAA0344, PRKWNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK1 (EC 2.7.11.1) (Protein kinase with no lysine 1) (Protein kinase, lysine-deficient 1) (Kinase deficient protein). | |||||
|
EGR2_MOUSE
|
||||||
| NC score | -0.000769 (rank : 49) | θ value | 8.99809 (rank : 42) | |||
| Query Neighborhood Hits | 49 | Target Neighborhood Hits | 745 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P08152 | Gene names | Egr2, Egr-2, Krox-20, Zfp-25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein). | |||||