Please be patient as the page loads
|
NUDT7_HUMAN
|
||||||
| SwissProt Accessions | P0C024 | Gene names | NUDT7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal coenzyme A diphosphatase NUDT7 (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 7) (Nudix motif 7). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NUDT7_HUMAN
|
||||||
| θ value | 1.33454e-136 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P0C024 | Gene names | NUDT7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal coenzyme A diphosphatase NUDT7 (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 7) (Nudix motif 7). | |||||
|
NUDT7_MOUSE
|
||||||
| θ value | 1.00358e-83 (rank : 2) | NC score | 0.985575 (rank : 2) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99P30, Q6IS65, Q8BU08, Q8K260, Q9D0Q3, Q9DBI9 | Gene names | Nudt7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal coenzyme A diphosphatase NUDT7 (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 7) (Nudix motif 7). | |||||
|
NUDT8_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 3) | NC score | 0.682507 (rank : 3) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WV74, Q6ZW59 | Gene names | NUDT8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 8, mitochondrial precursor (EC 3.6.1.-) (Nudix motif 8). | |||||
|
NUDT8_MOUSE
|
||||||
| θ value | 1.42992e-13 (rank : 4) | NC score | 0.678468 (rank : 4) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CR24, Q3TDQ2 | Gene names | Nudt8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 8, mitochondrial precursor (EC 3.6.1.-) (Nudix motif 8). | |||||
|
PGBM_MOUSE
|
||||||
| θ value | 1.38821 (rank : 5) | NC score | 0.010268 (rank : 6) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
GNAO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 6) | NC score | 0.010249 (rank : 7) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09471 | Gene names | GNAO1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(o) subunit alpha 1. | |||||
|
AP4A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 7) | NC score | 0.059128 (rank : 5) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50583 | Gene names | NUDT2, APAH1 | |||
|
Domain Architecture |
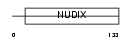
|
|||||
| Description | Bis(5'-nucleosyl)-tetraphosphatase [Asymmetrical] (EC 3.6.1.17) (Diadenosine 5',5'''-P1,P4-tetraphosphate asymmetrical hydrolase) (Diadenosine tetraphosphatase) (Ap4A hydrolase) (Ap4Aase) (Nucleoside diphosphate-linked moiety X motif 2) (Nudix motif 2). | |||||
|
NUDT7_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.33454e-136 (rank : 1) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P0C024 | Gene names | NUDT7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal coenzyme A diphosphatase NUDT7 (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 7) (Nudix motif 7). | |||||
|
NUDT7_MOUSE
|
||||||
| NC score | 0.985575 (rank : 2) | θ value | 1.00358e-83 (rank : 2) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99P30, Q6IS65, Q8BU08, Q8K260, Q9D0Q3, Q9DBI9 | Gene names | Nudt7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal coenzyme A diphosphatase NUDT7 (EC 3.6.1.-) (Nucleoside diphosphate-linked moiety X motif 7) (Nudix motif 7). | |||||
|
NUDT8_HUMAN
|
||||||
| NC score | 0.682507 (rank : 3) | θ value | 3.40345e-15 (rank : 3) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WV74, Q6ZW59 | Gene names | NUDT8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 8, mitochondrial precursor (EC 3.6.1.-) (Nudix motif 8). | |||||
|
NUDT8_MOUSE
|
||||||
| NC score | 0.678468 (rank : 4) | θ value | 1.42992e-13 (rank : 4) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9CR24, Q3TDQ2 | Gene names | Nudt8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoside diphosphate-linked moiety X motif 8, mitochondrial precursor (EC 3.6.1.-) (Nudix motif 8). | |||||
|
AP4A_HUMAN
|
||||||
| NC score | 0.059128 (rank : 5) | θ value | 4.03905 (rank : 7) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P50583 | Gene names | NUDT2, APAH1 | |||
|
Domain Architecture |

|
|||||
| Description | Bis(5'-nucleosyl)-tetraphosphatase [Asymmetrical] (EC 3.6.1.17) (Diadenosine 5',5'''-P1,P4-tetraphosphate asymmetrical hydrolase) (Diadenosine tetraphosphatase) (Ap4A hydrolase) (Ap4Aase) (Nucleoside diphosphate-linked moiety X motif 2) (Nudix motif 2). | |||||
|
PGBM_MOUSE
|
||||||
| NC score | 0.010268 (rank : 6) | θ value | 1.38821 (rank : 5) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 1073 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q05793 | Gene names | Hspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
GNAO1_HUMAN
|
||||||
| NC score | 0.010249 (rank : 7) | θ value | 3.0926 (rank : 6) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 75 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P09471 | Gene names | GNAO1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide-binding protein G(o) subunit alpha 1. | |||||