Please be patient as the page loads
|
NEU2_HUMAN
|
||||||
| SwissProt Accessions | P01185, O14935 | Gene names | AVP, ARVP, VP | |||
|
Domain Architecture |
|
|||||
| Description | Vasopressin-neurophysin 2-copeptin precursor (AVP-NPII) [Contains: Arg-vasopressin; Neurophysin 2 (Neurophysin-II); Copeptin]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NEU2_HUMAN
|
||||||
| θ value | 4.19693e-98 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P01185, O14935 | Gene names | AVP, ARVP, VP | |||
|
Domain Architecture |

|
|||||
| Description | Vasopressin-neurophysin 2-copeptin precursor (AVP-NPII) [Contains: Arg-vasopressin; Neurophysin 2 (Neurophysin-II); Copeptin]. | |||||
|
NEU2_MOUSE
|
||||||
| θ value | 2.82823e-78 (rank : 2) | NC score | 0.987915 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35455 | Gene names | Avp | |||
|
Domain Architecture |
|
|||||
| Description | Vasopressin-neurophysin 2-copeptin precursor (AVP-NPII) [Contains: Arg-vasopressin; Neurophysin 2 (Neurophysin-I); Copeptin]. | |||||
|
NEU1_MOUSE
|
||||||
| θ value | 1.08223e-53 (rank : 3) | NC score | 0.969034 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35454 | Gene names | Oxt | |||
|
Domain Architecture |
|
|||||
| Description | Oxytocin-neurophysin 1 precursor (OT-NPI) [Contains: Oxytocin (Ocytocin); Neurophysin 1]. | |||||
|
NEU1_HUMAN
|
||||||
| θ value | 2.66562e-52 (rank : 4) | NC score | 0.978417 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P01178, Q3MIG0 | Gene names | OXT, OT | |||
|
Domain Architecture |
|
|||||
| Description | Oxytocin-neurophysin 1 precursor (OT-NPI) [Contains: Oxytocin (Ocytocin); Neurophysin 1]. | |||||
|
KR106_HUMAN
|
||||||
| θ value | 0.21417 (rank : 5) | NC score | 0.066487 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR104_HUMAN
|
||||||
| θ value | 0.279714 (rank : 6) | NC score | 0.063577 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
TIE1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 7) | NC score | 0.024400 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1266 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q06806, Q811F4 | Gene names | Tie1, Tie, Tie-1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor Tie-1 precursor (EC 2.7.10.1). | |||||
|
TENX_HUMAN
|
||||||
| θ value | 0.47712 (rank : 8) | NC score | 0.046333 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
KRA58_HUMAN
|
||||||
| θ value | 0.62314 (rank : 9) | NC score | 0.079050 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
ERBB3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 10) | NC score | 0.022134 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P21860 | Gene names | ERBB3, HER3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Tyrosine kinase-type cell surface receptor HER3). | |||||
|
KRA55_HUMAN
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.079465 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
NID2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.029857 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
LRP2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.043570 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
TMPS6_MOUSE
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.015391 (rank : 41) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DBI0, Q6PF94 | Gene names | Tmprss6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
ADA11_HUMAN
|
||||||
| θ value | 2.36792 (rank : 15) | NC score | 0.021443 (rank : 38) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75078, Q14808, Q14809, Q14810 | Gene names | ADAM11, MDC | |||
|
Domain Architecture |
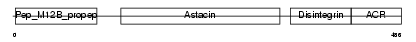
|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
ADA11_MOUSE
|
||||||
| θ value | 2.36792 (rank : 16) | NC score | 0.021431 (rank : 39) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9R1V4 | Gene names | Adam11, Mdc | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
PGBM_HUMAN
|
||||||
| θ value | 2.36792 (rank : 17) | NC score | 0.031495 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
TIE1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 18) | NC score | 0.022436 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P35590 | Gene names | TIE1, TIE | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor Tie-1 precursor (EC 2.7.10.1). | |||||
|
TMPS6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 19) | NC score | 0.015248 (rank : 42) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IU80, Q5TI06, Q6UXD8, Q8IUE2, Q8IXV8 | Gene names | TMPRSS6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
KR107_HUMAN
|
||||||
| θ value | 4.03905 (rank : 20) | NC score | 0.057041 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KR10C_HUMAN
|
||||||
| θ value | 4.03905 (rank : 21) | NC score | 0.072761 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
KRA51_HUMAN
|
||||||
| θ value | 4.03905 (rank : 22) | NC score | 0.061983 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
LRP1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 23) | NC score | 0.036516 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
MEGF8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 24) | NC score | 0.040443 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
MEGF8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 25) | NC score | 0.038046 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
NELL2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 26) | NC score | 0.033739 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61220 | Gene names | Nell2, Mel91 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (MEL91 protein). | |||||
|
STAB2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.037435 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
JAG2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 28) | NC score | 0.032924 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
JAG2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 29) | NC score | 0.035126 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
LRP8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 30) | NC score | 0.036440 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14114, O14968, Q86V27, Q99876, Q9BR78 | Gene names | LRP8, APOER2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
ERBB3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 31) | NC score | 0.016141 (rank : 40) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 898 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61526, Q3KQR1, Q68J64, Q810U8, Q8K317 | Gene names | Erbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Glial growth factor receptor). | |||||
|
KR108_HUMAN
|
||||||
| θ value | 6.88961 (rank : 32) | NC score | 0.051461 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
KRA54_HUMAN
|
||||||
| θ value | 6.88961 (rank : 33) | NC score | 0.040013 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
AGRIN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.033496 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
GRN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.040266 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P28798 | Gene names | Grn | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) (PC cell-derived growth factor) (PCDGF) [Contains: Acrogranin; Granulin-1; Granulin-2; Granulin-3; Granulin-4; Granulin-5; Granulin-6; Granulin-7]. | |||||
|
NELL2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.032226 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99435 | Gene names | NELL2, NRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (Nel-related protein 2). | |||||
|
RET_MOUSE
|
||||||
| θ value | 8.99809 (rank : 37) | NC score | 0.007428 (rank : 43) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35546, Q8BQ34, Q9QXH9 | Gene names | Ret | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase receptor ret precursor (EC 2.7.10.1) (C-ret). | |||||
|
KR103_HUMAN
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.051900 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
KR105_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.053905 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
KR171_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.077043 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
LCE2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.107661 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
LCE2C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.095336 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
LCE2D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.135249 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
NEU2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.19693e-98 (rank : 1) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P01185, O14935 | Gene names | AVP, ARVP, VP | |||
|
Domain Architecture |

|
|||||
| Description | Vasopressin-neurophysin 2-copeptin precursor (AVP-NPII) [Contains: Arg-vasopressin; Neurophysin 2 (Neurophysin-II); Copeptin]. | |||||
|
NEU2_MOUSE
|
||||||
| NC score | 0.987915 (rank : 2) | θ value | 2.82823e-78 (rank : 2) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35455 | Gene names | Avp | |||
|
Domain Architecture |

|
|||||
| Description | Vasopressin-neurophysin 2-copeptin precursor (AVP-NPII) [Contains: Arg-vasopressin; Neurophysin 2 (Neurophysin-I); Copeptin]. | |||||
|
NEU1_HUMAN
|
||||||
| NC score | 0.978417 (rank : 3) | θ value | 2.66562e-52 (rank : 4) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P01178, Q3MIG0 | Gene names | OXT, OT | |||
|
Domain Architecture |

|
|||||
| Description | Oxytocin-neurophysin 1 precursor (OT-NPI) [Contains: Oxytocin (Ocytocin); Neurophysin 1]. | |||||
|
NEU1_MOUSE
|
||||||
| NC score | 0.969034 (rank : 4) | θ value | 1.08223e-53 (rank : 3) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35454 | Gene names | Oxt | |||
|
Domain Architecture |

|
|||||
| Description | Oxytocin-neurophysin 1 precursor (OT-NPI) [Contains: Oxytocin (Ocytocin); Neurophysin 1]. | |||||
|
LCE2D_HUMAN
|
||||||
| NC score | 0.135249 (rank : 5) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5TA82 | Gene names | LCE2D, LEP12, SPRL1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2D (Late envelope protein 12) (Small proline-rich-like epidermal differentiation complex protein 1A). | |||||
|
LCE2B_HUMAN
|
||||||
| NC score | 0.107661 (rank : 6) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O14633 | Gene names | LCE2B, LEP10, SPRL1B, XP5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2B (Late envelope protein 10) (Small proline-rich-like epidermal differentiation complex protein 1B) (Skin- specific protein Xp5). | |||||
|
LCE2C_HUMAN
|
||||||
| NC score | 0.095336 (rank : 7) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5TA81 | Gene names | LCE2C, LEP11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Late cornified envelope protein 2C (Late envelope protein 11). | |||||
|
KRA55_HUMAN
|
||||||
| NC score | 0.079465 (rank : 8) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q701N2 | Gene names | KRTAP5-5, KAP5-11, KAP5.5, KRTAP5-11, KRTAP5.11, KRTAP5.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-5 (Keratin-associated protein 5.5) (Ultrahigh sulfur keratin-associated protein 5.5) (Keratin-associated protein 5-11) (Keratin-associated protein 5.11). | |||||
|
KRA58_HUMAN
|
||||||
| NC score | 0.079050 (rank : 9) | θ value | 0.62314 (rank : 9) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O75690, Q6L8G7, Q6UTX6 | Gene names | KRTAP5-8, KAP5.8, KRTAP5.8, UHSKB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-8 (Keratin-associated protein 5.8) (Ultrahigh sulfur keratin-associated protein 5.8) (Keratin, ultra high-sulfur matrix protein B) (UHS keratin B) (UHS KerB). | |||||
|
KR171_HUMAN
|
||||||
| NC score | 0.077043 (rank : 10) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 144 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BYP8 | Gene names | KRTAP17-1, KAP17.1, KRTAP16.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 17-1 (Keratin-associated protein 16.1). | |||||
|
KR10C_HUMAN
|
||||||
| NC score | 0.072761 (rank : 11) | θ value | 4.03905 (rank : 21) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P60413 | Gene names | KRTAP10-12, KAP10.12, KAP18-12, KRTAP10.12, KRTAP18-12, KRTAP18.12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-12 (Keratin-associated protein 10.12) (High sulfur keratin-associated protein 10.12) (Keratin-associated protein 18-12) (Keratin-associated protein 18.12). | |||||
|
KR106_HUMAN
|
||||||
| NC score | 0.066487 (rank : 12) | θ value | 0.21417 (rank : 5) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60371 | Gene names | KRTAP10-6, KAP10.6, KAP18-6, KRTAP10.6, KRTAP18-6, KRTAP18.6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-6 (Keratin-associated protein 10.6) (High sulfur keratin-associated protein 10.6) (Keratin-associated protein 18-6) (Keratin-associated protein 18.6). | |||||
|
KR104_HUMAN
|
||||||
| NC score | 0.063577 (rank : 13) | θ value | 0.279714 (rank : 6) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P60372 | Gene names | KRTAP10-4, KAP10.4, KAP18-4, KRTAP10.4, KRTAP18-4, KRTAP18.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-4 (Keratin-associated protein 10.4) (High sulfur keratin-associated protein 10.4) (Keratin-associated protein 18-4) (Keratin-associated protein 18.4). | |||||
|
KRA51_HUMAN
|
||||||
| NC score | 0.061983 (rank : 14) | θ value | 4.03905 (rank : 22) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6L8H4 | Gene names | KRTAP5-1, KAP5.1, KRTAP5.1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-1 (Keratin-associated protein 5.1) (Ultrahigh sulfur keratin-associated protein 5.1). | |||||
|
KR107_HUMAN
|
||||||
| NC score | 0.057041 (rank : 15) | θ value | 4.03905 (rank : 20) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P60409, Q70LJ2 | Gene names | KRTAP10-7, KAP10.7, KAP18-7, KRTAP10.7, KRTAP18-7, KRTAP18.7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-7 (Keratin-associated protein 10.7) (High sulfur keratin-associated protein 10.7) (Keratin-associated protein 18-7) (Keratin-associated protein 18.7). | |||||
|
KR105_HUMAN
|
||||||
| NC score | 0.053905 (rank : 16) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P60370, Q70LJ3 | Gene names | KRTAP10-5, KAP10.5, KAP18-5, KRTAP10.5, KRTAP18-5, KRTAP18.5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-5 (Keratin-associated protein 10.5) (High sulfur keratin-associated protein 10.5) (Keratin-associated protein 18-5) (Keratin-associated protein 18.5). | |||||
|
KR103_HUMAN
|
||||||
| NC score | 0.051900 (rank : 17) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P60369, Q70LJ4 | Gene names | KRTAP10-3, KAP10.3, KAP18-3, KRTAP10.3, KRTAP18-3, KRTAP18.3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-3 (Keratin-associated protein 10.3) (High sulfur keratin-associated protein 10.3) (Keratin-associated protein 18-3) (Keratin-associated protein 18.3). | |||||
|
KR108_HUMAN
|
||||||
| NC score | 0.051461 (rank : 18) | θ value | 6.88961 (rank : 32) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P60410 | Gene names | KRTAP10-8, KAP10.8, KAP18-8, KRTAP10.8, KRTAP18-8, KRTAP18.8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 10-8 (Keratin-associated protein 10.8) (High sulfur keratin-associated protein 10.8) (Keratin-associated protein 18-8) (Keratin-associated protein 18.8). | |||||
|
TENX_HUMAN
|
||||||
| NC score | 0.046333 (rank : 19) | θ value | 0.47712 (rank : 8) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 789 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P22105, P78530, P78531, Q08424, Q9UMG7 | Gene names | TNXB, HXBL, TNX, XB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin-X precursor (TN-X) (Hexabrachion-like protein). | |||||
|
LRP2_HUMAN
|
||||||
| NC score | 0.043570 (rank : 20) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 573 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P98164, O00711, Q16215 | Gene names | LRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 2 precursor (Megalin) (Glycoprotein 330) (gp330). | |||||
|
MEGF8_HUMAN
|
||||||
| NC score | 0.040443 (rank : 21) | θ value | 4.03905 (rank : 24) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 528 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q7Z7M0, O75097 | Gene names | MEGF8, EGFL4, KIAA0817 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
GRN_MOUSE
|
||||||
| NC score | 0.040266 (rank : 22) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P28798 | Gene names | Grn | |||
|
Domain Architecture |

|
|||||
| Description | Granulins precursor (Proepithelin) (PEPI) (PC cell-derived growth factor) (PCDGF) [Contains: Acrogranin; Granulin-1; Granulin-2; Granulin-3; Granulin-4; Granulin-5; Granulin-6; Granulin-7]. | |||||
|
KRA54_HUMAN
|
||||||
| NC score | 0.040013 (rank : 23) | θ value | 6.88961 (rank : 33) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q6L8H1 | Gene names | KRTAP5-4, KAP5.4, KRTAP5.4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin-associated protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh sulfur keratin-associated protein 5.4). | |||||
|
MEGF8_MOUSE
|
||||||
| NC score | 0.038046 (rank : 24) | θ value | 4.03905 (rank : 25) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 520 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P60882, Q80TR3, Q80V41, Q8BMN9, Q8JZW7, Q8K0J3 | Gene names | Megf8, Egfl4 | |||
|
Domain Architecture |

|
|||||
| Description | Multiple epidermal growth factor-like domains 8 (EGF-like domain- containing protein 4) (Multiple EGF-like domain protein 4). | |||||
|
STAB2_HUMAN
|
||||||
| NC score | 0.037435 (rank : 25) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 534 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8WWQ8, Q6ZMK2, Q7Z5N9, Q86UR4, Q8IUG9, Q8TES1, Q9H7H7, Q9NRY3 | Gene names | STAB2, FEEL2, FELL, FEX2, HARE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stabilin-2 precursor (FEEL-2 protein) (Fasciclin EGF-like laminin-type EGF-like and link domain-containing scavenger receptor 1) (FAS1 EGF- like and X-link domain-containing adhesion molecule 2) (Hyaluronan receptor for endocytosis) [Contains: 190 kDa form stabilin-2 (190 kDa hyaluronan receptor for endocytosis)]. | |||||
|
LRP1B_HUMAN
|
||||||
| NC score | 0.036516 (rank : 26) | θ value | 4.03905 (rank : 23) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 602 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NZR2, Q8WY29, Q8WY30, Q8WY31 | Gene names | LRP1B, LRPDIT | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 1B precursor (Low- density lipoprotein receptor-related protein-deleted in tumor) (LRP- DIT). | |||||
|
LRP8_HUMAN
|
||||||
| NC score | 0.036440 (rank : 27) | θ value | 5.27518 (rank : 30) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q14114, O14968, Q86V27, Q99876, Q9BR78 | Gene names | LRP8, APOER2 | |||
|
Domain Architecture |

|
|||||
| Description | Low-density lipoprotein receptor-related protein 8 precursor (Apolipoprotein E receptor 2). | |||||
|
JAG2_MOUSE
|
||||||
| NC score | 0.035126 (rank : 28) | θ value | 5.27518 (rank : 29) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 504 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9QYE5, O55139, O70219 | Gene names | Jag2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2). | |||||
|
NELL2_MOUSE
|
||||||
| NC score | 0.033739 (rank : 29) | θ value | 4.03905 (rank : 26) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q61220 | Gene names | Nell2, Mel91 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (MEL91 protein). | |||||
|
AGRIN_HUMAN
|
||||||
| NC score | 0.033496 (rank : 30) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 582 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O00468, Q5SVA2, Q60FE1, Q7KYS8, Q8N4J5, Q96IC1, Q9BTD4 | Gene names | AGRIN, AGRN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Agrin precursor. | |||||
|
JAG2_HUMAN
|
||||||
| NC score | 0.032924 (rank : 31) | θ value | 5.27518 (rank : 28) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9Y219, Q9UE17, Q9UE99, Q9UNK8, Q9Y6P9, Q9Y6Q0 | Gene names | JAG2 | |||
|
Domain Architecture |

|
|||||
| Description | Jagged-2 precursor (Jagged2) (HJ2). | |||||
|
NELL2_HUMAN
|
||||||
| NC score | 0.032226 (rank : 32) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 449 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99435 | Gene names | NELL2, NRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein NELL2 precursor (NEL-like protein 2) (Nel-related protein 2). | |||||
|
PGBM_HUMAN
|
||||||
| NC score | 0.031495 (rank : 33) | θ value | 2.36792 (rank : 17) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1113 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P98160, Q16287, Q9H3V5 | Gene names | HSPG2 | |||
|
Domain Architecture |

|
|||||
| Description | Basement membrane-specific heparan sulfate proteoglycan core protein precursor (HSPG) (Perlecan) (PLC). | |||||
|
NID2_HUMAN
|
||||||
| NC score | 0.029857 (rank : 34) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q14112, O43710 | Gene names | NID2 | |||
|
Domain Architecture |

|
|||||
| Description | Nidogen-2 precursor (NID-2) (Osteonidogen). | |||||
|
TIE1_MOUSE
|
||||||
| NC score | 0.024400 (rank : 35) | θ value | 0.279714 (rank : 7) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1266 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q06806, Q811F4 | Gene names | Tie1, Tie, Tie-1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor Tie-1 precursor (EC 2.7.10.1). | |||||
|
TIE1_HUMAN
|
||||||
| NC score | 0.022436 (rank : 36) | θ value | 3.0926 (rank : 18) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P35590 | Gene names | TIE1, TIE | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein kinase receptor Tie-1 precursor (EC 2.7.10.1). | |||||
|
ERBB3_HUMAN
|
||||||
| NC score | 0.022134 (rank : 37) | θ value | 1.06291 (rank : 10) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P21860 | Gene names | ERBB3, HER3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Tyrosine kinase-type cell surface receptor HER3). | |||||
|
ADA11_HUMAN
|
||||||
| NC score | 0.021443 (rank : 38) | θ value | 2.36792 (rank : 15) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 259 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O75078, Q14808, Q14809, Q14810 | Gene names | ADAM11, MDC | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
ADA11_MOUSE
|
||||||
| NC score | 0.021431 (rank : 39) | θ value | 2.36792 (rank : 16) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9R1V4 | Gene names | Adam11, Mdc | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 11 precursor (A disintegrin and metalloproteinase domain 11) (Metalloproteinase-like, disintegrin-like, and cysteine-rich protein) (MDC). | |||||
|
ERBB3_MOUSE
|
||||||
| NC score | 0.016141 (rank : 40) | θ value | 6.88961 (rank : 31) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 898 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q61526, Q3KQR1, Q68J64, Q810U8, Q8K317 | Gene names | Erbb3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Glial growth factor receptor). | |||||
|
TMPS6_MOUSE
|
||||||
| NC score | 0.015391 (rank : 41) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DBI0, Q6PF94 | Gene names | Tmprss6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
TMPS6_HUMAN
|
||||||
| NC score | 0.015248 (rank : 42) | θ value | 3.0926 (rank : 19) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IU80, Q5TI06, Q6UXD8, Q8IUE2, Q8IXV8 | Gene names | TMPRSS6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
RET_MOUSE
|
||||||
| NC score | 0.007428 (rank : 43) | θ value | 8.99809 (rank : 37) | |||
| Query Neighborhood Hits | 37 | Target Neighborhood Hits | 884 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35546, Q8BQ34, Q9QXH9 | Gene names | Ret | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase receptor ret precursor (EC 2.7.10.1) (C-ret). | |||||