Please be patient as the page loads
|
MTA70_HUMAN
|
||||||
| SwissProt Accessions | Q86U44, O14736, Q86V05, Q9HB32 | Gene names | METTL3, MTA70 | |||
|
Domain Architecture |

|
|||||
| Description | N6-adenosine-methyltransferase 70 kDa subunit (EC 2.1.1.62) (MT-A70) (Methyltransferase-like protein 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MTA70_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86U44, O14736, Q86V05, Q9HB32 | Gene names | METTL3, MTA70 | |||
|
Domain Architecture |

|
|||||
| Description | N6-adenosine-methyltransferase 70 kDa subunit (EC 2.1.1.62) (MT-A70) (Methyltransferase-like protein 3). | |||||
|
MTA70_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.995074 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C3P7, Q9CV54, Q9ERS9, Q9WUI4 | Gene names | Mettl3, Mta70 | |||
|
Domain Architecture |

|
|||||
| Description | N6-adenosine-methyltransferase 70 kDa subunit (EC 2.1.1.62) (MT-A70) (Methyltransferase-like protein 3). | |||||
|
METL4_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 3) | NC score | 0.357423 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3U034 | Gene names | Mettl4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyltransferase-like protein 4 (EC 2.1.1.-). | |||||
|
METL4_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 4) | NC score | 0.289941 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N3J2, Q2TAA7, Q9H5U9 | Gene names | METTL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyltransferase-like protein 4 (EC 2.1.1.-). | |||||
|
SDPR_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 5) | NC score | 0.106478 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 6) | NC score | 0.039442 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CO1A2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 7) | NC score | 0.027691 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01149, Q8CGA5 | Gene names | Col1a2, Cola2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
DKC1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 8) | NC score | 0.047480 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60832, O43845, Q96G67, Q9Y505 | Gene names | DKC1, NOLA4 | |||
|
Domain Architecture |
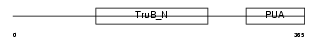
|
|||||
| Description | H/ACA ribonucleoprotein complex subunit 4 (EC 5.4.99.-) (Dyskerin) (Nucleolar protein family A member 4) (snoRNP protein DKC1) (Nopp140- associated protein of 57 kDa) (Nucleolar protein NAP57) (CBF5 homolog). | |||||
|
DNL1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 9) | NC score | 0.033897 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 10) | NC score | 0.021045 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
RBGPR_MOUSE
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.059748 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BMG7, Q6PEU0, Q80TQ6, Q8BX72 | Gene names | Rab3gap2, Kiaa0839 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab3 GTPase-activating protein non-catalytic subunit (Rab3 GTPase- activating protein 150 kDa subunit) (Rab3-GAP p150) (Rab3-GAP regulatory subunit) (RAB3-GAP150). | |||||
|
SC24A_MOUSE
|
||||||
| θ value | 1.06291 (rank : 12) | NC score | 0.030128 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
ATX2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 13) | NC score | 0.035017 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
IF4G1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.017150 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
ATX2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.030553 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.010877 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
MTF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.000851 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14872, Q96CB1 | Gene names | MTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||
|
SCMH1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.011854 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K214, Q9JME0 | Gene names | Scmh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SCMH1 (Sex comb on midleg homolog 1). | |||||
|
ULK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.001948 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
CIC_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.011780 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CIC_MOUSE
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.012570 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.009170 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
LAIR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.018014 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ISS4, Q6PEZ4 | Gene names | LAIR2, CD306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 2 precursor (LAIR-2) (CD306 antigen). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.007352 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 8.99809 (rank : 25) | NC score | 0.010062 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
SAPS2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 26) | NC score | 0.009386 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75170, Q9UGB9 | Gene names | SAPS2, KIAA0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
THSD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 27) | NC score | 0.017783 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NS62, Q6P3U1, Q6UXZ2 | Gene names | THSD1, TMTSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
MTA70_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q86U44, O14736, Q86V05, Q9HB32 | Gene names | METTL3, MTA70 | |||
|
Domain Architecture |

|
|||||
| Description | N6-adenosine-methyltransferase 70 kDa subunit (EC 2.1.1.62) (MT-A70) (Methyltransferase-like protein 3). | |||||
|
MTA70_MOUSE
|
||||||
| NC score | 0.995074 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8C3P7, Q9CV54, Q9ERS9, Q9WUI4 | Gene names | Mettl3, Mta70 | |||
|
Domain Architecture |

|
|||||
| Description | N6-adenosine-methyltransferase 70 kDa subunit (EC 2.1.1.62) (MT-A70) (Methyltransferase-like protein 3). | |||||
|
METL4_MOUSE
|
||||||
| NC score | 0.357423 (rank : 3) | θ value | 5.26297e-08 (rank : 3) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3U034 | Gene names | Mettl4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyltransferase-like protein 4 (EC 2.1.1.-). | |||||
|
METL4_HUMAN
|
||||||
| NC score | 0.289941 (rank : 4) | θ value | 0.00020696 (rank : 4) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N3J2, Q2TAA7, Q9H5U9 | Gene names | METTL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyltransferase-like protein 4 (EC 2.1.1.-). | |||||
|
SDPR_MOUSE
|
||||||
| NC score | 0.106478 (rank : 5) | θ value | 0.000786445 (rank : 5) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q63918, Q3V1P6, Q78EC3, Q8CBT4 | Gene names | Sdpr, Sdr | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serum deprivation-response protein (Phosphatidylserine-binding protein). | |||||
|
RBGPR_MOUSE
|
||||||
| NC score | 0.059748 (rank : 6) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BMG7, Q6PEU0, Q80TQ6, Q8BX72 | Gene names | Rab3gap2, Kiaa0839 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rab3 GTPase-activating protein non-catalytic subunit (Rab3 GTPase- activating protein 150 kDa subunit) (Rab3-GAP p150) (Rab3-GAP regulatory subunit) (RAB3-GAP150). | |||||
|
DKC1_HUMAN
|
||||||
| NC score | 0.047480 (rank : 7) | θ value | 0.62314 (rank : 8) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60832, O43845, Q96G67, Q9Y505 | Gene names | DKC1, NOLA4 | |||
|
Domain Architecture |

|
|||||
| Description | H/ACA ribonucleoprotein complex subunit 4 (EC 5.4.99.-) (Dyskerin) (Nucleolar protein family A member 4) (snoRNP protein DKC1) (Nopp140- associated protein of 57 kDa) (Nucleolar protein NAP57) (CBF5 homolog). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.039442 (rank : 8) | θ value | 0.0431538 (rank : 6) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
ATX2_HUMAN
|
||||||
| NC score | 0.035017 (rank : 9) | θ value | 1.81305 (rank : 13) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99700, Q6ZQZ7, Q99493 | Gene names | ATXN2, ATX2, SCA2, TNRC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein) (Trinucleotide repeat-containing gene 13 protein). | |||||
|
DNL1_HUMAN
|
||||||
| NC score | 0.033897 (rank : 10) | θ value | 0.813845 (rank : 9) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
ATX2_MOUSE
|
||||||
| NC score | 0.030553 (rank : 11) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O70305, P97421 | Gene names | Atxn2, Atx2, Sca2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2 (Spinocerebellar ataxia type 2 protein homolog). | |||||
|
SC24A_MOUSE
|
||||||
| NC score | 0.030128 (rank : 12) | θ value | 1.06291 (rank : 12) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 394 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3U2P1, Q3TQ05, Q3TRG7, Q8BIS0 | Gene names | Sec24a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein transport protein Sec24A (SEC24-related protein A). | |||||
|
CO1A2_MOUSE
|
||||||
| NC score | 0.027691 (rank : 13) | θ value | 0.279714 (rank : 7) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01149, Q8CGA5 | Gene names | Col1a2, Cola2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(I) chain precursor. | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.021045 (rank : 14) | θ value | 0.813845 (rank : 10) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
LAIR2_HUMAN
|
||||||
| NC score | 0.018014 (rank : 15) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 46 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ISS4, Q6PEZ4 | Gene names | LAIR2, CD306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte-associated immunoglobulin-like receptor 2 precursor (LAIR-2) (CD306 antigen). | |||||
|
THSD1_HUMAN
|
||||||
| NC score | 0.017783 (rank : 16) | θ value | 8.99809 (rank : 27) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NS62, Q6P3U1, Q6UXZ2 | Gene names | THSD1, TMTSP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thrombospondin type-1 domain-containing protein 1 precursor (Transmembrane molecule with thrombospondin module). | |||||
|
IF4G1_HUMAN
|
||||||
| NC score | 0.017150 (rank : 17) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q04637, O43177, O95066, Q5HYG0, Q6ZN21, Q8N102 | Gene names | EIF4G1, EIF4G, EIF4GI | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 4 gamma 1 (eIF-4-gamma 1) (eIF-4G1) (eIF-4G 1) (p220). | |||||
|
CIC_MOUSE
|
||||||
| NC score | 0.012570 (rank : 18) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 742 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q924A2, Q6PDJ8, Q8CGE4, Q8CHH0, Q9CW61 | Gene names | Cic, Kiaa0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
SCMH1_MOUSE
|
||||||
| NC score | 0.011854 (rank : 19) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K214, Q9JME0 | Gene names | Scmh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polycomb protein SCMH1 (Sex comb on midleg homolog 1). | |||||
|
CIC_HUMAN
|
||||||
| NC score | 0.011780 (rank : 20) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 992 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96RK0, Q7LGI1, Q9UEG5, Q9Y6T1 | Gene names | CIC, KIAA0306 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein capicua homolog. | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.010877 (rank : 21) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.010062 (rank : 22) | θ value | 8.99809 (rank : 25) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
SAPS2_HUMAN
|
||||||
| NC score | 0.009386 (rank : 23) | θ value | 8.99809 (rank : 26) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 41 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75170, Q9UGB9 | Gene names | SAPS2, KIAA0685 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 2. | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.009170 (rank : 24) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.007352 (rank : 25) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
ULK1_HUMAN
|
||||||
| NC score | 0.001948 (rank : 26) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 1121 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O75385 | Gene names | ULK1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase ULK1 (EC 2.7.11.1) (Unc-51-like kinase 1). | |||||
|
MTF1_HUMAN
|
||||||
| NC score | 0.000851 (rank : 27) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 27 | Target Neighborhood Hits | 814 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14872, Q96CB1 | Gene names | MTF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metal-regulatory transcription factor 1 (Transcription factor MTF-1) (MRE-binding transcription factor). | |||||