Please be patient as the page loads
|
LMA2L_MOUSE
|
||||||
| SwissProt Accessions | P59481, Q3TZ91 | Gene names | Lman2l, Vipl | |||
|
Domain Architecture |
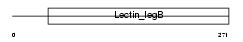
|
|||||
| Description | VIP36-like protein precursor (Lectin, mannose-binding 2-like) (LMAN2- like protein). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LMA2L_MOUSE
|
||||||
| θ value | 4.53388e-177 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59481, Q3TZ91 | Gene names | Lman2l, Vipl | |||
|
Domain Architecture |

|
|||||
| Description | VIP36-like protein precursor (Lectin, mannose-binding 2-like) (LMAN2- like protein). | |||||
|
LMA2L_HUMAN
|
||||||
| θ value | 1.56188e-169 (rank : 2) | NC score | 0.999590 (rank : 2) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H0V9, Q53GV3, Q53S67, Q63HN6, Q8NBQ6, Q9BQ14 | Gene names | LMAN2L, VIPL | |||
|
Domain Architecture |
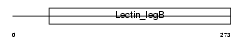
|
|||||
| Description | VIP36-like protein precursor (Lectin, mannose-binding 2-like) (LMAN2- like protein). | |||||
|
LMAN2_MOUSE
|
||||||
| θ value | 2.01279e-100 (rank : 3) | NC score | 0.986237 (rank : 4) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBH5, Q9CXG7 | Gene names | Lman2 | |||
|
Domain Architecture |
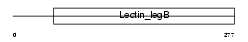
|
|||||
| Description | Vesicular integral-membrane protein VIP36 precursor (Lectin, mannose- binding 2). | |||||
|
LMAN2_HUMAN
|
||||||
| θ value | 1.70393e-99 (rank : 4) | NC score | 0.986363 (rank : 3) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12907, Q53HH1 | Gene names | LMAN2, C5orf8 | |||
|
Domain Architecture |
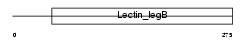
|
|||||
| Description | Vesicular integral-membrane protein VIP36 precursor (GP36b glycoprotein) (Lectin, mannose-binding 2). | |||||
|
LMAN1_MOUSE
|
||||||
| θ value | 1.42001e-37 (rank : 5) | NC score | 0.840766 (rank : 5) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D0F3 | Gene names | Lman1, Ergic53 | |||
|
Domain Architecture |
|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (p58). | |||||
|
LMAN1_HUMAN
|
||||||
| θ value | 1.01765e-35 (rank : 6) | NC score | 0.836992 (rank : 6) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49257, Q12895, Q8N5I7, Q9UQG1, Q9UQG2, Q9UQG3, Q9UQG4, Q9UQG5, Q9UQG6, Q9UQG7, Q9UQG8, Q9UQG9, Q9UQH0, Q9UQH1, Q9UQH2 | Gene names | LMAN1, ERGIC53, F5F8D | |||
|
Domain Architecture |
|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (Gp58) (Intracellular mannose- specific lectin MR60). | |||||
|
LMA1L_HUMAN
|
||||||
| θ value | 6.84181e-24 (rank : 7) | NC score | 0.806125 (rank : 8) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HAT1, Q6UWN2 | Gene names | LMAN1L, ERGL | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53-like protein precursor (ERGIC53-like protein) (Lectin, mannose-binding 1-like) (LMAN1-like protein). | |||||
|
LMA1L_MOUSE
|
||||||
| θ value | 2.59989e-23 (rank : 8) | NC score | 0.818303 (rank : 7) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VCD3 | Gene names | Lman1l, Ergl | |||
|
Domain Architecture |
|
|||||
| Description | ERGIC-53-like protein precursor (ERGIC53-like protein) (Lectin, mannose-binding 1-like) (LMAN1-like protein). | |||||
|
STAT6_MOUSE
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.033194 (rank : 9) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52633 | Gene names | Stat6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and transcription activator 6. | |||||
|
CJ064_HUMAN
|
||||||
| θ value | 8.99809 (rank : 10) | NC score | 0.011315 (rank : 10) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZS81, Q8N4A3 | Gene names | C10orf64 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf64. | |||||
|
PRAX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 11) | NC score | 0.007637 (rank : 11) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||
|
LMA2L_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.53388e-177 (rank : 1) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P59481, Q3TZ91 | Gene names | Lman2l, Vipl | |||
|
Domain Architecture |

|
|||||
| Description | VIP36-like protein precursor (Lectin, mannose-binding 2-like) (LMAN2- like protein). | |||||
|
LMA2L_HUMAN
|
||||||
| NC score | 0.999590 (rank : 2) | θ value | 1.56188e-169 (rank : 2) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H0V9, Q53GV3, Q53S67, Q63HN6, Q8NBQ6, Q9BQ14 | Gene names | LMAN2L, VIPL | |||
|
Domain Architecture |

|
|||||
| Description | VIP36-like protein precursor (Lectin, mannose-binding 2-like) (LMAN2- like protein). | |||||
|
LMAN2_HUMAN
|
||||||
| NC score | 0.986363 (rank : 3) | θ value | 1.70393e-99 (rank : 4) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q12907, Q53HH1 | Gene names | LMAN2, C5orf8 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicular integral-membrane protein VIP36 precursor (GP36b glycoprotein) (Lectin, mannose-binding 2). | |||||
|
LMAN2_MOUSE
|
||||||
| NC score | 0.986237 (rank : 4) | θ value | 2.01279e-100 (rank : 3) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DBH5, Q9CXG7 | Gene names | Lman2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicular integral-membrane protein VIP36 precursor (Lectin, mannose- binding 2). | |||||
|
LMAN1_MOUSE
|
||||||
| NC score | 0.840766 (rank : 5) | θ value | 1.42001e-37 (rank : 5) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9D0F3 | Gene names | Lman1, Ergic53 | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (p58). | |||||
|
LMAN1_HUMAN
|
||||||
| NC score | 0.836992 (rank : 6) | θ value | 1.01765e-35 (rank : 6) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P49257, Q12895, Q8N5I7, Q9UQG1, Q9UQG2, Q9UQG3, Q9UQG4, Q9UQG5, Q9UQG6, Q9UQG7, Q9UQG8, Q9UQG9, Q9UQH0, Q9UQH1, Q9UQH2 | Gene names | LMAN1, ERGIC53, F5F8D | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53 protein precursor (ER-Golgi intermediate compartment 53 kDa protein) (Lectin, mannose-binding 1) (Gp58) (Intracellular mannose- specific lectin MR60). | |||||
|
LMA1L_MOUSE
|
||||||
| NC score | 0.818303 (rank : 7) | θ value | 2.59989e-23 (rank : 8) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VCD3 | Gene names | Lman1l, Ergl | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53-like protein precursor (ERGIC53-like protein) (Lectin, mannose-binding 1-like) (LMAN1-like protein). | |||||
|
LMA1L_HUMAN
|
||||||
| NC score | 0.806125 (rank : 8) | θ value | 6.84181e-24 (rank : 7) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9HAT1, Q6UWN2 | Gene names | LMAN1L, ERGL | |||
|
Domain Architecture |

|
|||||
| Description | ERGIC-53-like protein precursor (ERGIC53-like protein) (Lectin, mannose-binding 1-like) (LMAN1-like protein). | |||||
|
STAT6_MOUSE
|
||||||
| NC score | 0.033194 (rank : 9) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52633 | Gene names | Stat6 | |||
|
Domain Architecture |

|
|||||
| Description | Signal transducer and transcription activator 6. | |||||
|
CJ064_HUMAN
|
||||||
| NC score | 0.011315 (rank : 10) | θ value | 8.99809 (rank : 10) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZS81, Q8N4A3 | Gene names | C10orf64 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C10orf64. | |||||
|
PRAX_HUMAN
|
||||||
| NC score | 0.007637 (rank : 11) | θ value | 8.99809 (rank : 11) | |||
| Query Neighborhood Hits | 11 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9BXM0, Q9BXL9, Q9HCF2 | Gene names | PRX, KIAA1620 | |||
|
Domain Architecture |

|
|||||
| Description | Periaxin. | |||||