Please be patient as the page loads
|
HSF4_MOUSE
|
||||||
| SwissProt Accessions | Q9R0L1, Q9R0L2 | Gene names | Hsf4 | |||
|
Domain Architecture |
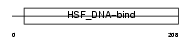
|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (mHSF4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
HSF4_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.984741 (rank : 2) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ULV5, Q99472, Q9ULV6 | Gene names | HSF4 | |||
|
Domain Architecture |
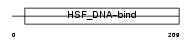
|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (hHSF4). | |||||
|
HSF4_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9R0L1, Q9R0L2 | Gene names | Hsf4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (mHSF4). | |||||
|
HSF1_HUMAN
|
||||||
| θ value | 4.67263e-73 (rank : 3) | NC score | 0.894422 (rank : 3) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
HSF1_MOUSE
|
||||||
| θ value | 4.83544e-70 (rank : 4) | NC score | 0.857710 (rank : 4) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
HSF2_MOUSE
|
||||||
| θ value | 2.94719e-51 (rank : 5) | NC score | 0.848473 (rank : 5) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P38533, Q64157, Q8VEJ0, Q9Z2L8 | Gene names | Hsf2 | |||
|
Domain Architecture |
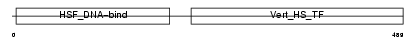
|
|||||
| Description | Heat shock factor protein 2 (HSF 2) (Heat shock transcription factor 2) (HSTF 2). | |||||
|
HSF2_HUMAN
|
||||||
| θ value | 8.57503e-51 (rank : 6) | NC score | 0.846823 (rank : 6) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03933, Q9H445 | Gene names | HSF2, HSTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 2 (HSF 2) (Heat shock transcription factor 2) (HSTF 2). | |||||
|
HSFY1_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 7) | NC score | 0.505072 (rank : 7) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96LI6, Q7Z4L8, Q9BZA2, Q9BZA3 | Gene names | HSFY1, HSF2L, HSFY | |||
|
Domain Architecture |
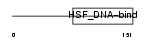
|
|||||
| Description | Heat shock transcription factor, Y-linked (Heat shock transcription factor 2-like protein) (HSF2-like). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 0.279714 (rank : 8) | NC score | 0.026544 (rank : 13) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.016793 (rank : 26) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
EPHA1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 10) | NC score | 0.003239 (rank : 55) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21709, Q15405 | Gene names | EPHA1, EPH, EPHT, EPHT1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 1 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EPH). | |||||
|
SYN2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 11) | NC score | 0.023224 (rank : 15) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92777 | Gene names | SYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
NFL_HUMAN
|
||||||
| θ value | 0.813845 (rank : 12) | NC score | 0.016826 (rank : 25) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
NFL_MOUSE
|
||||||
| θ value | 0.813845 (rank : 13) | NC score | 0.016672 (rank : 27) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
NKX28_HUMAN
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.010485 (rank : 42) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15522 | Gene names | NKX2-8, NKX-2.8, NKX2G, NKX2H | |||
|
Domain Architecture |
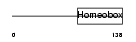
|
|||||
| Description | Homeobox protein Nkx-2.8 (Homeobox protein NK-2 homolog H). | |||||
|
PLCB4_HUMAN
|
||||||
| θ value | 0.813845 (rank : 15) | NC score | 0.022104 (rank : 16) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15147, Q5JYS8, Q5JYT0, Q5JYT4, Q9BQW5, Q9BQW6, Q9BQW8, Q9UJQ2 | Gene names | PLCB4 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 4 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-4) (PLC-beta-4). | |||||
|
GGN_MOUSE
|
||||||
| θ value | 1.06291 (rank : 16) | NC score | 0.031461 (rank : 11) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
ZN219_HUMAN
|
||||||
| θ value | 1.06291 (rank : 17) | NC score | 0.004761 (rank : 51) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2Y4, Q8IYC1, Q9BW28 | Gene names | ZNF219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 219. | |||||
|
MYO9B_MOUSE
|
||||||
| θ value | 1.38821 (rank : 18) | NC score | 0.010303 (rank : 43) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
TRI12_MOUSE
|
||||||
| θ value | 1.38821 (rank : 19) | NC score | 0.010122 (rank : 44) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PQ1, Q9D704 | Gene names | Trim12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 12. | |||||
|
E2F2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.017943 (rank : 23) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14209 | Gene names | E2F2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F2 (E2F-2). | |||||
|
WDR33_HUMAN
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.010551 (rank : 41) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
ATF5_HUMAN
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.047577 (rank : 8) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2D1, Q9BSA1, Q9UNQ3 | Gene names | ATF5, ATFX | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5) (Transcription factor ATFx). | |||||
|
CO4A1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.011222 (rank : 39) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
RSMB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.021790 (rank : 17) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14678, Q15490, Q9UIS5 | Gene names | SNRPB, COD, SNRPB1 | |||
|
Domain Architecture |
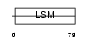
|
|||||
| Description | Small nuclear ribonucleoprotein-associated proteins B and B' (snRNP-B) (Sm protein B/B') (Sm-B/Sm-B') (SmB/SmB'). | |||||
|
ASXL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.020198 (rank : 18) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
CING_HUMAN
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.012640 (rank : 36) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
GNDS_HUMAN
|
||||||
| θ value | 3.0926 (rank : 27) | NC score | 0.019717 (rank : 19) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
HCN4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 28) | NC score | 0.008300 (rank : 48) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
MELPH_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.019661 (rank : 20) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
SPHK2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.019361 (rank : 21) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIA7, Q91VA9, Q9DBH6 | Gene names | Sphk2 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine kinase 2 (EC 2.7.1.-) (SK 2) (SPK 2). | |||||
|
CN004_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.035859 (rank : 10) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
SNX23_HUMAN
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.008620 (rank : 46) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
CN004_MOUSE
|
||||||
| θ value | 5.27518 (rank : 33) | NC score | 0.036517 (rank : 9) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
ZAR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.018715 (rank : 22) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
3BP2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 35) | NC score | 0.012696 (rank : 35) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
CO9A3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 36) | NC score | 0.008764 (rank : 45) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 37) | NC score | 0.017634 (rank : 24) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
GOSR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 38) | NC score | 0.025727 (rank : 14) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14653, Q96DA5, Q9BZZ4 | Gene names | GOSR2, GS27 | |||
|
Domain Architecture |
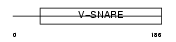
|
|||||
| Description | Golgi SNAP receptor complex member 2 (27 kDa Golgi SNARE protein) (Membrin). | |||||
|
GOSR2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.027198 (rank : 12) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35166, Q3UDN0, Q9CR77 | Gene names | Gosr2, Gs27 | |||
|
Domain Architecture |
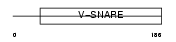
|
|||||
| Description | Golgi SNAP receptor complex member 2 (27 kDa Golgi SNARE protein) (Membrin). | |||||
|
IL4RA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.013986 (rank : 33) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
MRCKG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.001037 (rank : 57) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
MYPN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.004277 (rank : 52) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 418 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86TC9, Q5VV35, Q5VV36, Q86T37, Q8N3L4, Q96K90, Q96KF5 | Gene names | MYPN, MYOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin (145 kDa sarcomeric protein). | |||||
|
NEB2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.015463 (rank : 28) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
RNF5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.014698 (rank : 31) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99942, Q9UMQ2 | Gene names | RNF5, G16, NG2, RMA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase RMA1 (EC 6.3.2.-) (HsRma1) (RING finger protein 5) (Protein G16). | |||||
|
RNF5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.014859 (rank : 30) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35445 | Gene names | Rnf5, Ng2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase RMA1 (EC 6.3.2.-) (RING finger protein 5). | |||||
|
AINX_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.012415 (rank : 37) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16352, Q9BRC5 | Gene names | INA | |||
|
Domain Architecture |
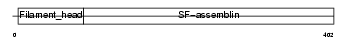
|
|||||
| Description | Alpha-internexin (Alpha-Inx) (66 kDa neurofilament protein) (Neurofilament-66) (NF-66). | |||||
|
CCHL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.013527 (rank : 34) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53701, Q502X8 | Gene names | HCCS, CCHL | |||
|
Domain Architecture |
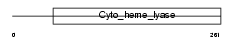
|
|||||
| Description | Cytochrome c-type heme lyase (EC 4.4.1.17) (CCHL) (Holocytochrome c- type synthase). | |||||
|
CE290_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.007827 (rank : 49) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CO4A3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.012243 (rank : 38) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
CO4A5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.015047 (rank : 29) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
IKBB_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.003860 (rank : 53) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15653, Q96BJ7 | Gene names | NFKBIB, IKBB, TRIP9 | |||
|
Domain Architecture |
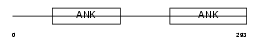
|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B) (Thyroid receptor-interacting protein 9) (TR-interacting protein 9). | |||||
|
MLL4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.010560 (rank : 40) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
NAT6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.014216 (rank : 32) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q93015, Q93014 | Gene names | NAT6, FUS2 | |||
|
Domain Architecture |
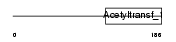
|
|||||
| Description | N-acetyltransferase 6 (EC 2.3.1.-) (Fus-2 protein) (Fusion 2 protein). | |||||
|
PTN18_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.003423 (rank : 54) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.007586 (rank : 50) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
TDRKH_HUMAN
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.008561 (rank : 47) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
ZSCA5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.001132 (rank : 56) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BUG6 | Gene names | ZSCAN5, ZNF495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 5 (Zinc finger protein 495). | |||||
|
HSF4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9R0L1, Q9R0L2 | Gene names | Hsf4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (mHSF4). | |||||
|
HSF4_HUMAN
|
||||||
| NC score | 0.984741 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9ULV5, Q99472, Q9ULV6 | Gene names | HSF4 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 4 (HSF 4) (Heat shock transcription factor 4) (HSTF 4) (hHSF4). | |||||
|
HSF1_HUMAN
|
||||||
| NC score | 0.894422 (rank : 3) | θ value | 4.67263e-73 (rank : 3) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q00613 | Gene names | HSF1, HSTF1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
HSF1_MOUSE
|
||||||
| NC score | 0.857710 (rank : 4) | θ value | 4.83544e-70 (rank : 4) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P38532, O70462 | Gene names | Hsf1 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 1 (HSF 1) (Heat shock transcription factor 1) (HSTF 1). | |||||
|
HSF2_MOUSE
|
||||||
| NC score | 0.848473 (rank : 5) | θ value | 2.94719e-51 (rank : 5) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P38533, Q64157, Q8VEJ0, Q9Z2L8 | Gene names | Hsf2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 2 (HSF 2) (Heat shock transcription factor 2) (HSTF 2). | |||||
|
HSF2_HUMAN
|
||||||
| NC score | 0.846823 (rank : 6) | θ value | 8.57503e-51 (rank : 6) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q03933, Q9H445 | Gene names | HSF2, HSTF2 | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock factor protein 2 (HSF 2) (Heat shock transcription factor 2) (HSTF 2). | |||||
|
HSFY1_HUMAN
|
||||||
| NC score | 0.505072 (rank : 7) | θ value | 4.92598e-06 (rank : 7) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96LI6, Q7Z4L8, Q9BZA2, Q9BZA3 | Gene names | HSFY1, HSF2L, HSFY | |||
|
Domain Architecture |

|
|||||
| Description | Heat shock transcription factor, Y-linked (Heat shock transcription factor 2-like protein) (HSF2-like). | |||||
|
ATF5_HUMAN
|
||||||
| NC score | 0.047577 (rank : 8) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Y2D1, Q9BSA1, Q9UNQ3 | Gene names | ATF5, ATFX | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-5 (Activating transcription factor 5) (Transcription factor ATFx). | |||||
|
CN004_MOUSE
|
||||||
| NC score | 0.036517 (rank : 9) | θ value | 5.27518 (rank : 33) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3X4, Q3USE5, Q8BUS1, Q8C011 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4 homolog. | |||||
|
CN004_HUMAN
|
||||||
| NC score | 0.035859 (rank : 10) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H1B7, Q8NDQ2, Q96JG2, Q9H3I7 | Gene names | C14orf4, KIAA1865 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf4. | |||||
|
GGN_MOUSE
|
||||||
| NC score | 0.031461 (rank : 11) | θ value | 1.06291 (rank : 16) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80WJ1, Q5EBP4, Q80WI9, Q80WJ0 | Gene names | Ggn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Gametogenetin. | |||||
|
GOSR2_MOUSE
|
||||||
| NC score | 0.027198 (rank : 12) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35166, Q3UDN0, Q9CR77 | Gene names | Gosr2, Gs27 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi SNAP receptor complex member 2 (27 kDa Golgi SNARE protein) (Membrin). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.026544 (rank : 13) | θ value | 0.279714 (rank : 8) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
GOSR2_HUMAN
|
||||||
| NC score | 0.025727 (rank : 14) | θ value | 6.88961 (rank : 38) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14653, Q96DA5, Q9BZZ4 | Gene names | GOSR2, GS27 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi SNAP receptor complex member 2 (27 kDa Golgi SNARE protein) (Membrin). | |||||
|
SYN2_HUMAN
|
||||||
| NC score | 0.023224 (rank : 15) | θ value | 0.62314 (rank : 11) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 213 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92777 | Gene names | SYN2 | |||
|
Domain Architecture |

|
|||||
| Description | Synapsin-2 (Synapsin II). | |||||
|
PLCB4_HUMAN
|
||||||
| NC score | 0.022104 (rank : 16) | θ value | 0.813845 (rank : 15) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 658 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q15147, Q5JYS8, Q5JYT0, Q5JYT4, Q9BQW5, Q9BQW6, Q9BQW8, Q9UJQ2 | Gene names | PLCB4 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 4 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-4) (PLC-beta-4). | |||||
|
RSMB_HUMAN
|
||||||
| NC score | 0.021790 (rank : 17) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14678, Q15490, Q9UIS5 | Gene names | SNRPB, COD, SNRPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Small nuclear ribonucleoprotein-associated proteins B and B' (snRNP-B) (Sm protein B/B') (Sm-B/Sm-B') (SmB/SmB'). | |||||
|
ASXL1_HUMAN
|
||||||
| NC score | 0.020198 (rank : 18) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IXJ9, Q5JWS9, Q8IYY7, Q9H466, Q9NQF8, Q9UFJ0, Q9UFP8, Q9Y2I4 | Gene names | ASXL1, KIAA0978 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative Polycomb group protein ASXL1 (Additional sex combs-like protein 1). | |||||
|
GNDS_HUMAN
|
||||||
| NC score | 0.019717 (rank : 19) | θ value | 3.0926 (rank : 27) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12967, Q9HAX7, Q9HAY1, Q9HCT1 | Gene names | RALGDS, RGF | |||
|
Domain Architecture |

|
|||||
| Description | Ral guanine nucleotide dissociation stimulator (RalGEF) (RalGDS). | |||||
|
MELPH_MOUSE
|
||||||
| NC score | 0.019661 (rank : 20) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91V27, Q99N53 | Gene names | Mlph, Ln, Slac2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Melanophilin (Exophilin-3) (Leaden protein) (Synaptotagmin-like protein 2a) (Slac2-a) (Slp homolog lacking C2 domains a). | |||||
|
SPHK2_MOUSE
|
||||||
| NC score | 0.019361 (rank : 21) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JIA7, Q91VA9, Q9DBH6 | Gene names | Sphk2 | |||
|
Domain Architecture |

|
|||||
| Description | Sphingosine kinase 2 (EC 2.7.1.-) (SK 2) (SPK 2). | |||||
|
ZAR1_HUMAN
|
||||||
| NC score | 0.018715 (rank : 22) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
E2F2_HUMAN
|
||||||
| NC score | 0.017943 (rank : 23) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14209 | Gene names | E2F2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription factor E2F2 (E2F-2). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.017634 (rank : 24) | θ value | 6.88961 (rank : 37) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
NFL_HUMAN
|
||||||
| NC score | 0.016826 (rank : 25) | θ value | 0.813845 (rank : 12) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 666 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P07196, Q16154, Q8IU72 | Gene names | NEFL, NF68, NFL | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.016793 (rank : 26) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
NFL_MOUSE
|
||||||
| NC score | 0.016672 (rank : 27) | θ value | 0.813845 (rank : 13) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 695 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P08551, Q8K0Z0 | Gene names | Nefl, Nf68, Nfl | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet L protein (68 kDa neurofilament protein) (Neurofilament light polypeptide) (NF-L). | |||||
|
NEB2_HUMAN
|
||||||
| NC score | 0.015463 (rank : 28) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 883 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96SB3, Q8TCR9 | Gene names | PPP1R9B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-2 (Neurabin-II) (Spinophilin) (Protein phosphatase 1 regulatory subunit 9B). | |||||
|
CO4A5_HUMAN
|
||||||
| NC score | 0.015047 (rank : 29) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P29400, Q16006, Q16126, Q6LD84 | Gene names | COL4A5 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-5(IV) chain precursor. | |||||
|
RNF5_MOUSE
|
||||||
| NC score | 0.014859 (rank : 30) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O35445 | Gene names | Rnf5, Ng2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase RMA1 (EC 6.3.2.-) (RING finger protein 5). | |||||
|
RNF5_HUMAN
|
||||||
| NC score | 0.014698 (rank : 31) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99942, Q9UMQ2 | Gene names | RNF5, G16, NG2, RMA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase RMA1 (EC 6.3.2.-) (HsRma1) (RING finger protein 5) (Protein G16). | |||||
|
NAT6_HUMAN
|
||||||
| NC score | 0.014216 (rank : 32) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q93015, Q93014 | Gene names | NAT6, FUS2 | |||
|
Domain Architecture |

|
|||||
| Description | N-acetyltransferase 6 (EC 2.3.1.-) (Fus-2 protein) (Fusion 2 protein). | |||||
|
IL4RA_MOUSE
|
||||||
| NC score | 0.013986 (rank : 33) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16382, O54690, Q60583, Q8CBW5 | Gene names | Il4r, Il4ra | |||
|
Domain Architecture |

|
|||||
| Description | Interleukin-4 receptor alpha chain precursor (IL-4R-alpha) (CD124 antigen) [Contains: Soluble interleukin-4 receptor alpha chain (IL-4- binding protein) (IL4-BP)]. | |||||
|
CCHL_HUMAN
|
||||||
| NC score | 0.013527 (rank : 34) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P53701, Q502X8 | Gene names | HCCS, CCHL | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome c-type heme lyase (EC 4.4.1.17) (CCHL) (Holocytochrome c- type synthase). | |||||
|
3BP2_MOUSE
|
||||||
| NC score | 0.012696 (rank : 35) | θ value | 6.88961 (rank : 35) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06649 | Gene names | Sh3bp2, 3bp2 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 2 (3BP-2). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.012640 (rank : 36) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
AINX_HUMAN
|
||||||
| NC score | 0.012415 (rank : 37) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 442 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q16352, Q9BRC5 | Gene names | INA | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-internexin (Alpha-Inx) (66 kDa neurofilament protein) (Neurofilament-66) (NF-66). | |||||
|
CO4A3_HUMAN
|
||||||
| NC score | 0.012243 (rank : 38) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
CO4A1_MOUSE
|
||||||
| NC score | 0.011222 (rank : 39) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 450 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P02463 | Gene names | Col4a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
MLL4_HUMAN
|
||||||
| NC score | 0.010560 (rank : 40) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UMN6, O15022, O95836, Q96GP2, Q96IP3, Q9UK25, Q9Y668, Q9Y669 | Gene names | MLL4, HRX2, KIAA0304, MLL2, TRX2 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloid/lymphoid or mixed-lineage leukemia protein 4 (Trithorax homolog 2). | |||||
|
WDR33_HUMAN
|
||||||
| NC score | 0.010551 (rank : 41) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 638 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9C0J8 | Gene names | WDR33, WDC146 | |||
|
Domain Architecture |

|
|||||
| Description | WD repeat protein 33 (WD repeat protein WDC146). | |||||
|
NKX28_HUMAN
|
||||||
| NC score | 0.010485 (rank : 42) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15522 | Gene names | NKX2-8, NKX-2.8, NKX2G, NKX2H | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Nkx-2.8 (Homeobox protein NK-2 homolog H). | |||||
|
MYO9B_MOUSE
|
||||||
| NC score | 0.010303 (rank : 43) | θ value | 1.38821 (rank : 18) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QY06, Q9QY07, Q9QY08, Q9QY09 | Gene names | Myo9b, Myr5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
TRI12_MOUSE
|
||||||
| NC score | 0.010122 (rank : 44) | θ value | 1.38821 (rank : 19) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 387 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99PQ1, Q9D704 | Gene names | Trim12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 12. | |||||
|
CO9A3_HUMAN
|
||||||
| NC score | 0.008764 (rank : 45) | θ value | 6.88961 (rank : 36) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
SNX23_HUMAN
|
||||||
| NC score | 0.008620 (rank : 46) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
TDRKH_HUMAN
|
||||||
| NC score | 0.008561 (rank : 47) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y2W6, Q8N582, Q9NYV5 | Gene names | TDRKH, TDRD2 | |||
|
Domain Architecture |

|
|||||
| Description | Tudor and KH domain-containing protein. | |||||
|
HCN4_MOUSE
|
||||||
| NC score | 0.008300 (rank : 48) | θ value | 3.0926 (rank : 28) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O70507 | Gene names | Hcn4, Bcng3 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 (Brain cyclic nucleotide-gated channel 3) (BCNG-3). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.007827 (rank : 49) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.007586 (rank : 50) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
ZN219_HUMAN
|
||||||
| NC score | 0.004761 (rank : 51) | θ value | 1.06291 (rank : 17) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 876 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P2Y4, Q8IYC1, Q9BW28 | Gene names | ZNF219 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 219. | |||||
|
MYPN_HUMAN
|
||||||
| NC score | 0.004277 (rank : 52) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 418 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q86TC9, Q5VV35, Q5VV36, Q86T37, Q8N3L4, Q96K90, Q96KF5 | Gene names | MYPN, MYOP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myopalladin (145 kDa sarcomeric protein). | |||||
|
IKBB_HUMAN
|
||||||
| NC score | 0.003860 (rank : 53) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15653, Q96BJ7 | Gene names | NFKBIB, IKBB, TRIP9 | |||
|
Domain Architecture |

|
|||||
| Description | NF-kappa-B inhibitor beta (NF-kappa-BIB) (I-kappa-B-beta) (IkappaBbeta) (IKB-beta) (IKB-B) (Thyroid receptor-interacting protein 9) (TR-interacting protein 9). | |||||
|
PTN18_MOUSE
|
||||||
| NC score | 0.003423 (rank : 54) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
EPHA1_HUMAN
|
||||||
| NC score | 0.003239 (rank : 55) | θ value | 0.62314 (rank : 10) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 997 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P21709, Q15405 | Gene names | EPHA1, EPH, EPHT, EPHT1 | |||
|
Domain Architecture |

|
|||||
| Description | Ephrin type-A receptor 1 precursor (EC 2.7.10.1) (Tyrosine-protein kinase receptor EPH). | |||||
|
ZSCA5_HUMAN
|
||||||
| NC score | 0.001132 (rank : 56) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 862 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BUG6 | Gene names | ZSCAN5, ZNF495 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger and SCAN domain-containing protein 5 (Zinc finger protein 495). | |||||
|
MRCKG_HUMAN
|
||||||
| NC score | 0.001037 (rank : 57) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||