Please be patient as the page loads
|
GDAP1_HUMAN
|
||||||
| SwissProt Accessions | Q8TB36 | Gene names | GDAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ganglioside-induced differentiation-associated protein 1 (GDAP1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GDAP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8TB36 | Gene names | GDAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ganglioside-induced differentiation-associated protein 1 (GDAP1). | |||||
|
GDAP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998148 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O88741, Q8C7Q5, Q9CTN2 | Gene names | Gdap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ganglioside-induced differentiation-associated protein 1 (GDAP1). | |||||
|
GD1L1_HUMAN
|
||||||
| θ value | 5.46864e-106 (rank : 3) | NC score | 0.958317 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96MZ0, Q5TE60, Q68CW7, Q9BQJ7, Q9BQV4, Q9BWJ4, Q9H3Y2, Q9H4G5 | Gene names | GDAP1L1 | |||
|
Domain Architecture |
|
|||||
| Description | Ganglioside-induced differentiation-associated protein 1-like 1 (GDAP1-L1). | |||||
|
GD1L1_MOUSE
|
||||||
| θ value | 3.91911e-104 (rank : 4) | NC score | 0.956015 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VE33 | Gene names | Gdap1l1, Gdap1l | |||
|
Domain Architecture |
|
|||||
| Description | Ganglioside-induced differentiation-associated protein 1-like 1 (GDAP1-L1). | |||||
|
MAAI_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 5) | NC score | 0.149277 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43708, O15308, O75430, Q7Z610, Q9BV63 | Gene names | GSTZ1, MAAI | |||
|
Domain Architecture |
|
|||||
| Description | Maleylacetoacetate isomerase (EC 5.2.1.2) (MAAI) (Glutathione S- transferase zeta 1) (EC 2.5.1.18) (GSTZ1-1). | |||||
|
CLIC5_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 6) | NC score | 0.094065 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BXK9, Q3U0H8 | Gene names | Clic5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 5. | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 7) | NC score | 0.065235 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
TXD11_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.072294 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PKC3, O95887, Q6PJA6, Q8N2Q4, Q96K45, Q96K53 | Gene names | TXNDC11, EFP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11 (EF-hand-binding protein 1). | |||||
|
GSTO1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 9) | NC score | 0.119161 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09131, Q3TH87 | Gene names | Gsto1, Gstx, Gtsttl | |||
|
Domain Architecture |
|
|||||
| Description | Glutathione transferase omega-1 (EC 2.5.1.18) (GSTO 1-1) (p28). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 0.125558 (rank : 10) | NC score | 0.053658 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
GSTT2_HUMAN
|
||||||
| θ value | 0.125558 (rank : 11) | NC score | 0.107321 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P30712, O60665, Q9HD76 | Gene names | GSTT2 | |||
|
Domain Architecture |
|
|||||
| Description | Glutathione S-transferase theta-2 (EC 2.5.1.18) (GST class-theta-2). | |||||
|
GSTO1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.121742 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P78417, Q5TA03, Q7Z3T2 | Gene names | GSTO1, GSTTLP28 | |||
|
Domain Architecture |
|
|||||
| Description | Glutathione transferase omega-1 (EC 2.5.1.18) (GSTO 1-1). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.060756 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
LUZP1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.067794 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
CLIC5_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.085928 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZA1, Q5T4Z0, Q96JT5, Q9BWZ0 | Gene names | CLIC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 5. | |||||
|
MAAI_MOUSE
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.128694 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WVL0 | Gene names | Gstz1, Maai | |||
|
Domain Architecture |
|
|||||
| Description | Maleylacetoacetate isomerase (EC 5.2.1.2) (MAAI) (Glutathione S- transferase zeta 1) (EC 2.5.1.18) (GSTZ1-1). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.048145 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 18) | NC score | 0.046390 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
GSTT1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 19) | NC score | 0.086109 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64471 | Gene names | Gstt1 | |||
|
Domain Architecture |
|
|||||
| Description | Glutathione S-transferase theta-1 (EC 2.5.1.18) (GST class-theta-1). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.037295 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
UACA_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.040692 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
K2C1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.024574 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||
|
MYO6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.028850 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
CLIC6_MOUSE
|
||||||
| θ value | 1.81305 (rank : 24) | NC score | 0.063031 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
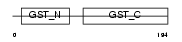
GSTT2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 25) | NC score | 0.081924 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61133, Q61134, Q64472 | Gene names | Gstt2 | |||
|
Domain Architecture |
|
|||||
| Description | Glutathione S-transferase theta-2 (EC 2.5.1.18) (GST class-theta-2). | |||||
|
NUF2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.050377 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
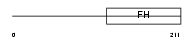
FOXJ1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.009202 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61660 | Gene names | Foxj1, Fkhl13, Hfh4 | |||
|
Domain Architecture |
|
|||||
| Description | Forkhead box protein J1 (Forkhead-related protein FKHL13) (Hepatocyte nuclear factor 3 forkhead homolog 4) (HFH-4). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.041822 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 2.36792 (rank : 29) | NC score | 0.041873 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 30) | NC score | 0.045432 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.043730 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
C102B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 32) | NC score | 0.053274 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q68D86, Q7Z467, Q8NDK7, Q9H5C1 | Gene names | CCDC102B, C18orf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 102B. | |||||
|
CLIC2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 33) | NC score | 0.068949 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15247, O15174, Q5JT80, Q8TCE3 | Gene names | CLIC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 2 (XAP121). | |||||
|
CLIC4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 34) | NC score | 0.070094 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QYB1, Q8BMG5 | Gene names | Clic4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 4 (mc3s5/mtCLIC). | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.054273 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
CLIC4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 36) | NC score | 0.067592 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y696, Q9UFW9, Q9UQJ6 | Gene names | CLIC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 4 (Intracellular chloride ion channel protein p64H1). | |||||
|
EXOC1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 37) | NC score | 0.037272 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NV70, Q8WUE7, Q96T15, Q9NZE4 | Gene names | EXOC1, SEC3, SEC3L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 1 (Exocyst complex component Sec3). | |||||
|
STOX1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 38) | NC score | 0.027553 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZVD7, Q4F8Q6, Q5I946, Q5I947, Q5I948, Q5VX38, Q5VX39, Q6ZRY3, Q96LR3, Q96LS0 | Gene names | STOX1, C10orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Storkhead-box protein 1 (Winged helix domain-containing protein). | |||||
|
U2AFL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 39) | NC score | 0.026228 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.037839 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
EVPL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.038094 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
GCC1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.036332 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96CN9, Q9H6N7 | Gene names | GCC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
PLCB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 43) | NC score | 0.026015 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
S23IP_MOUSE
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.017701 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.037874 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.018241 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.019040 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
DNMT1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.023564 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P13864, P97413, Q80ZU3, Q9CSC6, Q9QXX6 | Gene names | Dnmt1, Dnmt, Met1, Uim | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase MmuI) (DNA MTase MmuI) (MCMT) (M.MmuI) (Met-1). | |||||
|
DYNA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.036882 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
DYNA_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.037502 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
LAMA2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.020286 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
MYH13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.035137 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 53) | NC score | 0.033209 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 54) | NC score | 0.033731 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
POSTN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.015547 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15063, Q15064, Q5VSY5, Q8IZF9 | Gene names | POSTN, OSF2 | |||
|
Domain Architecture |

|
|||||
| Description | Periostin precursor (PN) (Osteoblast-specific factor 2) (OSF-2). | |||||
|
TPM3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.035248 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.037656 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
DMD_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.033822 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 59) | NC score | 0.039627 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 60) | NC score | 0.039324 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 61) | NC score | 0.035713 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.036806 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
LRRF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.025840 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.036500 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
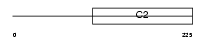
RFIP5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.014823 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXF6, O94939, Q9P0M1 | Gene names | RAB11FIP5, GAF1, KIAA0857, RIP11 | |||
|
Domain Architecture |
|
|||||
| Description | Rab11 family-interacting protein 5 (Rab11-FIP5) (Rab11-interacting protein Rip11) (Gamma-SNAP-associated factor 1) (Gaf-1) (Phosphoprotein pp75). | |||||
|
RNBP6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.008776 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60518, Q5T7X4, Q7Z3V2, Q96E78 | Gene names | RANBP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 6 (RanBP6). | |||||
|
CLIC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.050084 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00299, Q15089 | Gene names | CLIC1, NCC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 1 (Nuclear chloride ion channel 27) (NCC27) (Chloride channel ABP) (Regulatory nuclear chloride ion channel protein) (hRNCC). | |||||
|
CLIC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.050161 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z1Q5 | Gene names | Clic1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 1 (Nuclear chloride ion channel 27) (NCC27). | |||||
|
CLIC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.053167 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D7P7 | Gene names | Clic3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 3. | |||||
|
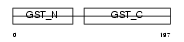
GSTT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.050803 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30711, O00226, Q969K8, Q96IY3 | Gene names | GSTT1 | |||
|
Domain Architecture |
|
|||||
| Description | Glutathione S-transferase theta-1 (EC 2.5.1.18) (GST class-theta-1) (Glutathione transferase T1-1). | |||||
|
GDAP1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 66 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8TB36 | Gene names | GDAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ganglioside-induced differentiation-associated protein 1 (GDAP1). | |||||
|
GDAP1_MOUSE
|
||||||
| NC score | 0.998148 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 64 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | O88741, Q8C7Q5, Q9CTN2 | Gene names | Gdap1 | |||
|
Domain Architecture |

|
|||||
| Description | Ganglioside-induced differentiation-associated protein 1 (GDAP1). | |||||
|
GD1L1_HUMAN
|
||||||
| NC score | 0.958317 (rank : 3) | θ value | 5.46864e-106 (rank : 3) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96MZ0, Q5TE60, Q68CW7, Q9BQJ7, Q9BQV4, Q9BWJ4, Q9H3Y2, Q9H4G5 | Gene names | GDAP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Ganglioside-induced differentiation-associated protein 1-like 1 (GDAP1-L1). | |||||
|
GD1L1_MOUSE
|
||||||
| NC score | 0.956015 (rank : 4) | θ value | 3.91911e-104 (rank : 4) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VE33 | Gene names | Gdap1l1, Gdap1l | |||
|
Domain Architecture |

|
|||||
| Description | Ganglioside-induced differentiation-associated protein 1-like 1 (GDAP1-L1). | |||||
|
MAAI_HUMAN
|
||||||
| NC score | 0.149277 (rank : 5) | θ value | 0.0330416 (rank : 5) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O43708, O15308, O75430, Q7Z610, Q9BV63 | Gene names | GSTZ1, MAAI | |||
|
Domain Architecture |

|
|||||
| Description | Maleylacetoacetate isomerase (EC 5.2.1.2) (MAAI) (Glutathione S- transferase zeta 1) (EC 2.5.1.18) (GSTZ1-1). | |||||
|
MAAI_MOUSE
|
||||||
| NC score | 0.128694 (rank : 6) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WVL0 | Gene names | Gstz1, Maai | |||
|
Domain Architecture |

|
|||||
| Description | Maleylacetoacetate isomerase (EC 5.2.1.2) (MAAI) (Glutathione S- transferase zeta 1) (EC 2.5.1.18) (GSTZ1-1). | |||||
|
GSTO1_HUMAN
|
||||||
| NC score | 0.121742 (rank : 7) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P78417, Q5TA03, Q7Z3T2 | Gene names | GSTO1, GSTTLP28 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione transferase omega-1 (EC 2.5.1.18) (GSTO 1-1). | |||||
|
GSTO1_MOUSE
|
||||||
| NC score | 0.119161 (rank : 8) | θ value | 0.0961366 (rank : 9) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O09131, Q3TH87 | Gene names | Gsto1, Gstx, Gtsttl | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione transferase omega-1 (EC 2.5.1.18) (GSTO 1-1) (p28). | |||||
|
GSTT2_HUMAN
|
||||||
| NC score | 0.107321 (rank : 9) | θ value | 0.125558 (rank : 11) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P30712, O60665, Q9HD76 | Gene names | GSTT2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase theta-2 (EC 2.5.1.18) (GST class-theta-2). | |||||
|
CLIC5_MOUSE
|
||||||
| NC score | 0.094065 (rank : 10) | θ value | 0.0431538 (rank : 6) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BXK9, Q3U0H8 | Gene names | Clic5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 5. | |||||
|
GSTT1_MOUSE
|
||||||
| NC score | 0.086109 (rank : 11) | θ value | 0.813845 (rank : 19) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64471 | Gene names | Gstt1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase theta-1 (EC 2.5.1.18) (GST class-theta-1). | |||||
|
CLIC5_HUMAN
|
||||||
| NC score | 0.085928 (rank : 12) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9NZA1, Q5T4Z0, Q96JT5, Q9BWZ0 | Gene names | CLIC5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 5. | |||||
|
GSTT2_MOUSE
|
||||||
| NC score | 0.081924 (rank : 13) | θ value | 1.81305 (rank : 25) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q61133, Q61134, Q64472 | Gene names | Gstt2 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase theta-2 (EC 2.5.1.18) (GST class-theta-2). | |||||
|
TXD11_HUMAN
|
||||||
| NC score | 0.072294 (rank : 14) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q6PKC3, O95887, Q6PJA6, Q8N2Q4, Q96K45, Q96K53 | Gene names | TXNDC11, EFP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 11 (EF-hand-binding protein 1). | |||||
|
CLIC4_MOUSE
|
||||||
| NC score | 0.070094 (rank : 15) | θ value | 3.0926 (rank : 34) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9QYB1, Q8BMG5 | Gene names | Clic4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 4 (mc3s5/mtCLIC). | |||||
|
CLIC2_HUMAN
|
||||||
| NC score | 0.068949 (rank : 16) | θ value | 3.0926 (rank : 33) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O15247, O15174, Q5JT80, Q8TCE3 | Gene names | CLIC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 2 (XAP121). | |||||
|
LUZP1_HUMAN
|
||||||
| NC score | 0.067794 (rank : 17) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 739 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q86V48, Q5TH93, Q8N4X3, Q8TEH1 | Gene names | LUZP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1. | |||||
|
CLIC4_HUMAN
|
||||||
| NC score | 0.067592 (rank : 18) | θ value | 4.03905 (rank : 36) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9Y696, Q9UFW9, Q9UQJ6 | Gene names | CLIC4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 4 (Intracellular chloride ion channel protein p64H1). | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.065235 (rank : 19) | θ value | 0.0563607 (rank : 7) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
CLIC6_MOUSE
|
||||||
| NC score | 0.063031 (rank : 20) | θ value | 1.81305 (rank : 24) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.060756 (rank : 21) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.054273 (rank : 22) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.053658 (rank : 23) | θ value | 0.125558 (rank : 10) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
C102B_HUMAN
|
||||||
| NC score | 0.053274 (rank : 24) | θ value | 3.0926 (rank : 32) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q68D86, Q7Z467, Q8NDK7, Q9H5C1 | Gene names | CCDC102B, C18orf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 102B. | |||||
|
CLIC3_MOUSE
|
||||||
| NC score | 0.053167 (rank : 25) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9D7P7 | Gene names | Clic3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 3. | |||||
|
GSTT1_HUMAN
|
||||||
| NC score | 0.050803 (rank : 26) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30711, O00226, Q969K8, Q96IY3 | Gene names | GSTT1 | |||
|
Domain Architecture |

|
|||||
| Description | Glutathione S-transferase theta-1 (EC 2.5.1.18) (GST class-theta-1) (Glutathione transferase T1-1). | |||||
|
NUF2_HUMAN
|
||||||
| NC score | 0.050377 (rank : 27) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
CLIC1_MOUSE
|
||||||
| NC score | 0.050161 (rank : 28) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Z1Q5 | Gene names | Clic1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 1 (Nuclear chloride ion channel 27) (NCC27). | |||||
|
CLIC1_HUMAN
|
||||||
| NC score | 0.050084 (rank : 29) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O00299, Q15089 | Gene names | CLIC1, NCC27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel protein 1 (Nuclear chloride ion channel 27) (NCC27) (Chloride channel ABP) (Regulatory nuclear chloride ion channel protein) (hRNCC). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.048145 (rank : 30) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.046390 (rank : 31) | θ value | 0.813845 (rank : 18) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.045432 (rank : 32) | θ value | 2.36792 (rank : 30) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.043730 (rank : 33) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.041873 (rank : 34) | θ value | 2.36792 (rank : 29) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.041822 (rank : 35) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.040692 (rank : 36) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.039627 (rank : 37) | θ value | 8.99809 (rank : 59) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.039324 (rank : 38) | θ value | 8.99809 (rank : 60) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
EVPL_MOUSE
|
||||||
| NC score | 0.038094 (rank : 39) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.037874 (rank : 40) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.037839 (rank : 41) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.037656 (rank : 42) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
DYNA_MOUSE
|
||||||
| NC score | 0.037502 (rank : 43) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O08788 | Gene names | Dctn1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.037295 (rank : 44) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
EXOC1_HUMAN
|
||||||
| NC score | 0.037272 (rank : 45) | θ value | 4.03905 (rank : 37) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9NV70, Q8WUE7, Q96T15, Q9NZE4 | Gene names | EXOC1, SEC3, SEC3L1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exocyst complex component 1 (Exocyst complex component Sec3). | |||||
|
DYNA_HUMAN
|
||||||
| NC score | 0.036882 (rank : 46) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1194 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q14203, O95296, Q9BRM9, Q9UIU1, Q9UIU2 | Gene names | DCTN1 | |||
|
Domain Architecture |

|
|||||
| Description | Dynactin-1 (150 kDa dynein-associated polypeptide) (DP-150) (DAP-150) (p150-glued) (p135). | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.036806 (rank : 47) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.036500 (rank : 48) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
GCC1_HUMAN
|
||||||
| NC score | 0.036332 (rank : 49) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 887 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96CN9, Q9H6N7 | Gene names | GCC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP and coiled-coil domain-containing protein 1 (Golgi coiled coil protein 1). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.035713 (rank : 50) | θ value | 8.99809 (rank : 61) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
TPM3_HUMAN
|
||||||
| NC score | 0.035248 (rank : 51) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 689 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P06753, P12324, Q969Q2, Q9NQH8 | Gene names | TPM3 | |||
|
Domain Architecture |

|
|||||
| Description | Tropomyosin alpha-3 chain (Tropomyosin-3) (Tropomyosin gamma) (hTM5). | |||||
|
MYH13_HUMAN
|
||||||
| NC score | 0.035137 (rank : 52) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1504 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q9UKX3, O95252 | Gene names | MYH13 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-13 (Myosin heavy chain, skeletal muscle, extraocular) (MyHC- eo). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.033822 (rank : 53) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.033731 (rank : 54) | θ value | 6.88961 (rank : 54) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.033209 (rank : 55) | θ value | 6.88961 (rank : 53) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
MYO6_MOUSE
|
||||||
| NC score | 0.028850 (rank : 56) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 586 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q64331 | Gene names | Myo6, Sv | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
STOX1_HUMAN
|
||||||
| NC score | 0.027553 (rank : 57) | θ value | 4.03905 (rank : 38) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 43 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q6ZVD7, Q4F8Q6, Q5I946, Q5I947, Q5I948, Q5VX38, Q5VX39, Q6ZRY3, Q96LR3, Q96LS0 | Gene names | STOX1, C10orf24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Storkhead-box protein 1 (Winged helix domain-containing protein). | |||||
|
U2AFL_HUMAN
|
||||||
| NC score | 0.026228 (rank : 58) | θ value | 4.03905 (rank : 39) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
PLCB1_HUMAN
|
||||||
| NC score | 0.026015 (rank : 59) | θ value | 5.27518 (rank : 43) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
LRRF1_MOUSE
|
||||||
| NC score | 0.025840 (rank : 60) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
K2C1_HUMAN
|
||||||
| NC score | 0.024574 (rank : 61) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P04264, Q14720, Q6GSJ0, Q9H298 | Gene names | KRT1, KRTA | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 1 (Cytokeratin-1) (CK-1) (Keratin-1) (K1) (67 kDa cytokeratin) (Hair alpha protein). | |||||
|
DNMT1_MOUSE
|
||||||
| NC score | 0.023564 (rank : 62) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P13864, P97413, Q80ZU3, Q9CSC6, Q9QXX6 | Gene names | Dnmt1, Dnmt, Met1, Uim | |||
|
Domain Architecture |

|
|||||
| Description | DNA (cytosine-5)-methyltransferase 1 (EC 2.1.1.37) (Dnmt1) (DNA methyltransferase MmuI) (DNA MTase MmuI) (MCMT) (M.MmuI) (Met-1). | |||||
|
LAMA2_HUMAN
|
||||||
| NC score | 0.020286 (rank : 63) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P24043, Q14736, Q93022 | Gene names | LAMA2, LAMM | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-2 chain precursor (Laminin M chain) (Merosin heavy chain). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.019040 (rank : 64) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.018241 (rank : 65) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
S23IP_MOUSE
|
||||||
| NC score | 0.017701 (rank : 66) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q6NZC7, Q8BXY6 | Gene names | Sec23ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SEC23-interacting protein. | |||||
|
POSTN_HUMAN
|
||||||
| NC score | 0.015547 (rank : 67) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15063, Q15064, Q5VSY5, Q8IZF9 | Gene names | POSTN, OSF2 | |||
|
Domain Architecture |

|
|||||
| Description | Periostin precursor (PN) (Osteoblast-specific factor 2) (OSF-2). | |||||
|
RFIP5_HUMAN
|
||||||
| NC score | 0.014823 (rank : 68) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BXF6, O94939, Q9P0M1 | Gene names | RAB11FIP5, GAF1, KIAA0857, RIP11 | |||
|
Domain Architecture |

|
|||||
| Description | Rab11 family-interacting protein 5 (Rab11-FIP5) (Rab11-interacting protein Rip11) (Gamma-SNAP-associated factor 1) (Gaf-1) (Phosphoprotein pp75). | |||||
|
FOXJ1_MOUSE
|
||||||
| NC score | 0.009202 (rank : 69) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61660 | Gene names | Foxj1, Fkhl13, Hfh4 | |||
|
Domain Architecture |

|
|||||
| Description | Forkhead box protein J1 (Forkhead-related protein FKHL13) (Hepatocyte nuclear factor 3 forkhead homolog 4) (HFH-4). | |||||
|
RNBP6_HUMAN
|
||||||
| NC score | 0.008776 (rank : 70) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 66 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60518, Q5T7X4, Q7Z3V2, Q96E78 | Gene names | RANBP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 6 (RanBP6). | |||||