Please be patient as the page loads
|
GAS2_MOUSE
|
||||||
| SwissProt Accessions | P11862 | Gene names | Gas2, Gas-2 | |||
|
Domain Architecture |
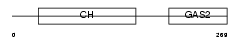
|
|||||
| Description | Growth-arrest-specific protein 2 (GAS-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GAS2_MOUSE
|
||||||
| θ value | 2.85304e-163 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P11862 | Gene names | Gas2, Gas-2 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 2 (GAS-2). | |||||
|
GAS2_HUMAN
|
||||||
| θ value | 4.26331e-159 (rank : 2) | NC score | 0.999567 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43903, Q6ICV8 | Gene names | GAS2 | |||
|
Domain Architecture |
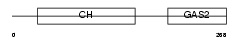
|
|||||
| Description | Growth-arrest-specific protein 2 (GAS-2). | |||||
|
GA2L3_HUMAN
|
||||||
| θ value | 5.88354e-84 (rank : 3) | NC score | 0.899565 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
GA2L2_HUMAN
|
||||||
| θ value | 8.04462e-41 (rank : 4) | NC score | 0.845690 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
GA2L2_MOUSE
|
||||||
| θ value | 8.04462e-41 (rank : 5) | NC score | 0.769778 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
GA2L1_HUMAN
|
||||||
| θ value | 3.06405e-32 (rank : 6) | NC score | 0.822218 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
GA2L1_MOUSE
|
||||||
| θ value | 6.82597e-32 (rank : 7) | NC score | 0.832377 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8JZP9, Q5SVG0, Q8K573 | Gene names | Gas2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1). | |||||
|
MACF1_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 8) | NC score | 0.209379 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 5.81887e-07 (rank : 9) | NC score | 0.203287 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 5.81887e-07 (rank : 10) | NC score | 0.223847 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 9.92553e-07 (rank : 11) | NC score | 0.188129 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BPAEA_HUMAN
|
||||||
| θ value | 1.29631e-06 (rank : 12) | NC score | 0.213655 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
LRCH4_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 13) | NC score | 0.068800 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q921G6 | Gene names | Lrch4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4. | |||||
|
LRCH4_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 14) | NC score | 0.068363 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75427, Q8WV85, Q96ID0 | Gene names | LRCH4, LRN, LRRN4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4 (Leucine-rich neuronal protein). | |||||
|
LRCH1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.043863 (rank : 46) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P62046 | Gene names | Lrch1, Chdc1, Kiaa1016 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 1 (Calponin homology domain-containing protein 1). | |||||
|
CNN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.048968 (rank : 45) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99439, Q92578 | Gene names | CNN2 | |||
|
Domain Architecture |
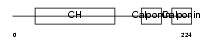
|
|||||
| Description | Calponin-2 (Calponin H2, smooth muscle) (Neutral calponin). | |||||
|
CNN1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.072067 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P51911, O00638, Q15416, Q8IY93, Q99438 | Gene names | CNN1 | |||
|
Domain Architecture |
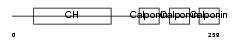
|
|||||
| Description | Calponin-1 (Calponin H1, smooth muscle) (Basic calponin). | |||||
|
CNN2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.049660 (rank : 44) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q08093 | Gene names | Cnn2 | |||
|
Domain Architecture |
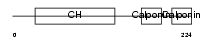
|
|||||
| Description | Calponin-2 (Calponin H2, smooth muscle) (Neutral calponin). | |||||
|
CNN1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.070452 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q08091 | Gene names | Cnn1 | |||
|
Domain Architecture |
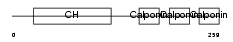
|
|||||
| Description | Calponin-1 (Calponin H1, smooth muscle) (Basic calponin). | |||||
|
DBF4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.020318 (rank : 49) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBU7, O75226, Q75MS6, Q75N01, Q9Y2M6 | Gene names | DBF4, ASK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DBF4 homolog (Activator of S phase Kinase). | |||||
|
LRCH3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.039833 (rank : 47) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96II8, Q96FP9, Q9NT52 | Gene names | LRCH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 3 precursor. | |||||
|
LRCH3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.037957 (rank : 48) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BVU0, Q3U222, Q3UZ74 | Gene names | Lrch3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 3 precursor. | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.077808 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CLMN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 24) | NC score | 0.050151 (rank : 42) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
CLMN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.050082 (rank : 43) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
CNN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.058455 (rank : 24) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DAW9, Q3TW23 | Gene names | Cnn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calponin-3 (Calponin, acidic isoform). | |||||
|
DESP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.058361 (rank : 27) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
DMD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.059495 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DMD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.058448 (rank : 25) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
DRP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.051043 (rank : 40) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13474 | Gene names | DRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin-related protein 2. | |||||
|
EVPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.050897 (rank : 41) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
EVPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.052548 (rank : 37) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
PEPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.052779 (rank : 35) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
PEPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.054787 (rank : 33) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.068378 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
SMOO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.051198 (rank : 39) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
SPTA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.056740 (rank : 29) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
SPTA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.061255 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
SPTA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.060108 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
SPTB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.055765 (rank : 31) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.055951 (rank : 30) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.053875 (rank : 34) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.052733 (rank : 36) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
SPTN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.058405 (rank : 26) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
SPTN4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.055371 (rank : 32) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.057642 (rank : 28) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.059309 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
SYNE2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.051981 (rank : 38) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
UTRO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.061248 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
GAS2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 2.85304e-163 (rank : 1) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P11862 | Gene names | Gas2, Gas-2 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 2 (GAS-2). | |||||
|
GAS2_HUMAN
|
||||||
| NC score | 0.999567 (rank : 2) | θ value | 4.26331e-159 (rank : 2) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43903, Q6ICV8 | Gene names | GAS2 | |||
|
Domain Architecture |

|
|||||
| Description | Growth-arrest-specific protein 2 (GAS-2). | |||||
|
GA2L3_HUMAN
|
||||||
| NC score | 0.899565 (rank : 3) | θ value | 5.88354e-84 (rank : 3) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86XJ1 | Gene names | GAS2L3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 3 (Growth arrest-specific 2-like 3). | |||||
|
GA2L2_HUMAN
|
||||||
| NC score | 0.845690 (rank : 4) | θ value | 8.04462e-41 (rank : 4) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8NHY3, Q8NHY4 | Gene names | GAS2L2, GAR17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2) (GAS2-related protein on chromosome 17) (GAR17 protein). | |||||
|
GA2L1_MOUSE
|
||||||
| NC score | 0.832377 (rank : 5) | θ value | 6.82597e-32 (rank : 7) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8JZP9, Q5SVG0, Q8K573 | Gene names | Gas2l1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1). | |||||
|
GA2L1_HUMAN
|
||||||
| NC score | 0.822218 (rank : 6) | θ value | 3.06405e-32 (rank : 6) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99501, Q53EN7, Q92640, Q9BUY9 | Gene names | GAS2L1, GAR22 | |||
|
Domain Architecture |

|
|||||
| Description | GAS2-like protein 1 (Growth arrest-specific 2-like 1) (GAS2-related protein on chromosome 22) (GAR22 protein). | |||||
|
GA2L2_MOUSE
|
||||||
| NC score | 0.769778 (rank : 7) | θ value | 8.04462e-41 (rank : 5) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q5SSG4 | Gene names | Gas2l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GAS2-like protein 2 (Growth arrest-specific 2-like 2). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.223847 (rank : 8) | θ value | 5.81887e-07 (rank : 10) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
BPAEA_HUMAN
|
||||||
| NC score | 0.213655 (rank : 9) | θ value | 1.29631e-06 (rank : 12) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1369 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O94833, Q5TBT0, Q8N1T8, Q8N8J3, Q8WXK9, Q96AK9, Q96DQ5, Q9H555 | Gene names | DST, BPAG1, DMH, DT, KIAA0728 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 6/9/10 (Trabeculin-beta) (Bullous pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MACF1_HUMAN
|
||||||
| NC score | 0.209379 (rank : 10) | θ value | 2.36244e-08 (rank : 8) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1341 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPN3, O75053, Q5VW20, Q8WXY2, Q9H540, Q9UKP0, Q9ULG9 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5 (Actin cross-linking family protein 7) (Macrophin-1) (Trabeculin-alpha) (620 kDa actin-binding protein) (ABP620). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.203287 (rank : 11) | θ value | 5.81887e-07 (rank : 9) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.188129 (rank : 12) | θ value | 9.92553e-07 (rank : 11) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.077808 (rank : 13) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CNN1_HUMAN
|
||||||
| NC score | 0.072067 (rank : 14) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P51911, O00638, Q15416, Q8IY93, Q99438 | Gene names | CNN1 | |||
|
Domain Architecture |

|
|||||
| Description | Calponin-1 (Calponin H1, smooth muscle) (Basic calponin). | |||||
|
CNN1_MOUSE
|
||||||
| NC score | 0.070452 (rank : 15) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q08091 | Gene names | Cnn1 | |||
|
Domain Architecture |

|
|||||
| Description | Calponin-1 (Calponin H1, smooth muscle) (Basic calponin). | |||||
|
LRCH4_MOUSE
|
||||||
| NC score | 0.068800 (rank : 16) | θ value | 0.0193708 (rank : 13) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q921G6 | Gene names | Lrch4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4. | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.068378 (rank : 17) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
LRCH4_HUMAN
|
||||||
| NC score | 0.068363 (rank : 18) | θ value | 0.0252991 (rank : 14) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O75427, Q8WV85, Q96ID0 | Gene names | LRCH4, LRN, LRRN4 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 4 (Leucine-rich neuronal protein). | |||||
|
SPTA1_MOUSE
|
||||||
| NC score | 0.061255 (rank : 19) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 675 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P08032, P97502 | Gene names | Spta1, Spna1, Spta | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
UTRO_HUMAN
|
||||||
| NC score | 0.061248 (rank : 20) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P46939 | Gene names | UTRN, DMDL | |||
|
Domain Architecture |

|
|||||
| Description | Utrophin (Dystrophin-related protein 1) (DRP1) (DRP). | |||||
|
SPTA2_HUMAN
|
||||||
| NC score | 0.060108 (rank : 21) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 888 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13813, Q13186, Q15324, Q16606, Q59EF1, Q5VXV5, Q5VXV6, Q7Z6M5, Q9P0V0 | Gene names | SPTAN1, SPTA2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, brain (Spectrin, non-erythroid alpha chain) (Alpha-II spectrin) (Fodrin alpha chain). | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.059495 (rank : 22) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.059309 (rank : 23) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
CNN3_MOUSE
|
||||||
| NC score | 0.058455 (rank : 24) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9DAW9, Q3TW23 | Gene names | Cnn3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calponin-3 (Calponin, acidic isoform). | |||||
|
DMD_MOUSE
|
||||||
| NC score | 0.058448 (rank : 25) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1111 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11531, O35653, Q60703 | Gene names | Dmd | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
SPTN2_HUMAN
|
||||||
| NC score | 0.058405 (rank : 26) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15020, O14872, O14873 | Gene names | SPTBN2, KIAA0302 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 2 (Spectrin, non-erythroid beta chain 2) (Beta-III spectrin). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.058361 (rank : 27) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.057642 (rank : 28) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
SPTA1_HUMAN
|
||||||
| NC score | 0.056740 (rank : 29) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02549, Q15514, Q6LDY5 | Gene names | SPTA1, SPTA | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin alpha chain, erythrocyte (Erythroid alpha-spectrin). | |||||
|
SPTB1_MOUSE
|
||||||
| NC score | 0.055951 (rank : 30) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTB1_HUMAN
|
||||||
| NC score | 0.055765 (rank : 31) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
SPTN4_HUMAN
|
||||||
| NC score | 0.055371 (rank : 32) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H254, Q9H1K7, Q9H1K8, Q9H1K9, Q9H3G8, Q9HCD0 | Gene names | SPTBN4, KIAA1642, SPTBN3 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 3 (Spectrin, non-erythroid beta chain 3) (Beta-IV spectrin). | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.054787 (rank : 33) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
SPTB2_HUMAN
|
||||||
| NC score | 0.053875 (rank : 34) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 748 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q01082, O60837, Q16057 | Gene names | SPTBN1, SPTB2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.052779 (rank : 35) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
SPTB2_MOUSE
|
||||||
| NC score | 0.052733 (rank : 36) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 770 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62261, Q5SQL8 | Gene names | Sptbn1, Spnb-2, Spnb2, Sptb2 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 1 (Spectrin, non-erythroid beta chain 1) (Beta-II spectrin) (Fodrin beta chain). | |||||
|
EVPL_MOUSE
|
||||||
| NC score | 0.052548 (rank : 37) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
SYNE2_HUMAN
|
||||||
| NC score | 0.051981 (rank : 38) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 1323 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8WXH0, Q8N1S3, Q8NF49, Q8TER7, Q8WWW3, Q8WWW4, Q8WWW5, Q8WXH1, Q9NU50, Q9UFQ4, Q9Y2L4, Q9Y4R1 | Gene names | SYNE2, KIAA1011, NUA | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-2 (Nuclear envelope spectrin repeat protein 2) (Syne-2) (Synaptic nuclear envelope protein 2) (Nucleus and actin connecting element protein) (Protein NUANCE). | |||||
|
SMOO_HUMAN
|
||||||
| NC score | 0.051198 (rank : 39) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P53814, O00569, O95769, O95937, Q9P1S8, Q9UIT1, Q9UIT2 | Gene names | SMTN, SMSMO | |||
|
Domain Architecture |

|
|||||
| Description | Smoothelin. | |||||
|
DRP2_HUMAN
|
||||||
| NC score | 0.051043 (rank : 40) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13474 | Gene names | DRP2 | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin-related protein 2. | |||||
|
EVPL_HUMAN
|
||||||
| NC score | 0.050897 (rank : 41) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
CLMN_HUMAN
|
||||||
| NC score | 0.050151 (rank : 42) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96JQ2, Q9H713, Q9HA23, Q9HA57, Q9UFP4, Q9ULN2 | Gene names | CLMN, KIAA1188 | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
CLMN_MOUSE
|
||||||
| NC score | 0.050082 (rank : 43) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8C5W0, Q91V71, Q91XT7, Q91XT8, Q91XU9 | Gene names | Clmn | |||
|
Domain Architecture |

|
|||||
| Description | Calmin. | |||||
|
CNN2_MOUSE
|
||||||
| NC score | 0.049660 (rank : 44) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q08093 | Gene names | Cnn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calponin-2 (Calponin H2, smooth muscle) (Neutral calponin). | |||||
|
CNN2_HUMAN
|
||||||
| NC score | 0.048968 (rank : 45) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 37 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99439, Q92578 | Gene names | CNN2 | |||
|
Domain Architecture |

|
|||||
| Description | Calponin-2 (Calponin H2, smooth muscle) (Neutral calponin). | |||||
|
LRCH1_MOUSE
|
||||||
| NC score | 0.043863 (rank : 46) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P62046 | Gene names | Lrch1, Chdc1, Kiaa1016 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 1 (Calponin homology domain-containing protein 1). | |||||
|
LRCH3_HUMAN
|
||||||
| NC score | 0.039833 (rank : 47) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 270 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96II8, Q96FP9, Q9NT52 | Gene names | LRCH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 3 precursor. | |||||
|
LRCH3_MOUSE
|
||||||
| NC score | 0.037957 (rank : 48) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 257 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8BVU0, Q3U222, Q3UZ74 | Gene names | Lrch3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeats and calponin homology domain-containing protein 3 precursor. | |||||
|
DBF4_HUMAN
|
||||||
| NC score | 0.020318 (rank : 49) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 22 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBU7, O75226, Q75MS6, Q75N01, Q9Y2M6 | Gene names | DBF4, ASK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DBF4 homolog (Activator of S phase Kinase). | |||||