Please be patient as the page loads
|
GABT_MOUSE
|
||||||
| SwissProt Accessions | P61922, Q8BZA3 | Gene names | Abat, Gabat | |||
|
Domain Architecture |

|
|||||
| Description | 4-aminobutyrate aminotransferase, mitochondrial precursor (EC 2.6.1.19) ((S)-3-amino-2-methylpropionate transaminase) (EC 2.6.1.22) (Gamma-amino-N-butyrate transaminase) (GABA transaminase) (GABA aminotransferase) (GABA-AT) (GABA-T) (L-AIBAT). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
GABT_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998673 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80404, Q16260, Q8N5W2, Q96BG2, Q99800 | Gene names | ABAT, GABAT | |||
|
Domain Architecture |

|
|||||
| Description | 4-aminobutyrate aminotransferase, mitochondrial precursor (EC 2.6.1.19) ((S)-3-amino-2-methylpropionate transaminase) (EC 2.6.1.22) (Gamma-amino-N-butyrate transaminase) (GABA transaminase) (GABA aminotransferase) (GABA-AT) (GABA-T) (L-AIBAT). | |||||
|
GABT_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P61922, Q8BZA3 | Gene names | Abat, Gabat | |||
|
Domain Architecture |

|
|||||
| Description | 4-aminobutyrate aminotransferase, mitochondrial precursor (EC 2.6.1.19) ((S)-3-amino-2-methylpropionate transaminase) (EC 2.6.1.22) (Gamma-amino-N-butyrate transaminase) (GABA transaminase) (GABA aminotransferase) (GABA-AT) (GABA-T) (L-AIBAT). | |||||
|
AGT2_HUMAN
|
||||||
| θ value | 4.91457e-14 (rank : 3) | NC score | 0.586737 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYV1, Q53FB4, Q53FY7, Q53G03, Q5W7Q1 | Gene names | AGXT2, AGT2 | |||
|
Domain Architecture |

|
|||||
| Description | Alanine--glyoxylate aminotransferase 2, mitochondrial precursor (EC 2.6.1.44) ((R)-3-amino-2-methylpropionate--pyruvate transaminase) (EC 2.6.1.40) (AGT 2) (Beta-alanine-pyruvate aminotransferase) (Beta- ALAAT II) (D-AIBAT). | |||||
|
OAT_MOUSE
|
||||||
| θ value | 1.58096e-12 (rank : 4) | NC score | 0.524575 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29758 | Gene names | Oat | |||
|
Domain Architecture |
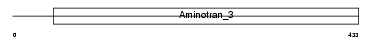
|
|||||
| Description | Ornithine aminotransferase, mitochondrial precursor (EC 2.6.1.13) (Ornithine--oxo-acid aminotransferase). | |||||
|
OAT_HUMAN
|
||||||
| θ value | 6.00763e-12 (rank : 5) | NC score | 0.520235 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P04181, Q16068, Q16069, Q9UD03 | Gene names | OAT | |||
|
Domain Architecture |
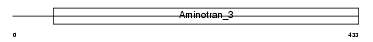
|
|||||
| Description | Ornithine aminotransferase, mitochondrial precursor (EC 2.6.1.13) (Ornithine--oxo-acid aminotransferase) [Contains: Ornithine aminotransferase, hepatic form; Ornithine aminotransferase, renal form]. | |||||
|
CDC23_MOUSE
|
||||||
| θ value | 3.0926 (rank : 6) | NC score | 0.035067 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGZ4, Q8C0F1 | Gene names | Cdc23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
CDC23_HUMAN
|
||||||
| θ value | 4.03905 (rank : 7) | NC score | 0.033571 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJX2, O75433, Q9BS73 | Gene names | CDC23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
UB2Q2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 8) | NC score | 0.022787 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WVN8, Q8N4G6, Q96J08 | Gene names | UBE2Q2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 Q2 (EC 6.3.2.19) (Ubiquitin-protein ligase Q2) (Ubiquitin carrier protein Q2). | |||||
|
ZN292_HUMAN
|
||||||
| θ value | 6.88961 (rank : 9) | NC score | 0.009939 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
ASB15_MOUSE
|
||||||
| θ value | 8.99809 (rank : 10) | NC score | 0.004333 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHS6 | Gene names | Asb15 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 15 (ASB-15). | |||||
|
PI52B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 11) | NC score | 0.022573 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78356, Q5U0E8, Q8TBP2 | Gene names | PIP5K2B | |||
|
Domain Architecture |
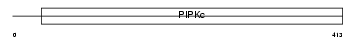
|
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-2 beta (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type II beta) (1- phosphatidylinositol-4-phosphate 5-kinase 2-beta) (PtdIns(4)P-5-kinase isoform 2-beta) (PIP5KII-beta) (Diphosphoinositide kinase 2-beta). | |||||
|
TM1L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 12) | NC score | 0.011032 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75674 | Gene names | TOM1L1, SRCASM | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
GABT_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P61922, Q8BZA3 | Gene names | Abat, Gabat | |||
|
Domain Architecture |

|
|||||
| Description | 4-aminobutyrate aminotransferase, mitochondrial precursor (EC 2.6.1.19) ((S)-3-amino-2-methylpropionate transaminase) (EC 2.6.1.22) (Gamma-amino-N-butyrate transaminase) (GABA transaminase) (GABA aminotransferase) (GABA-AT) (GABA-T) (L-AIBAT). | |||||
|
GABT_HUMAN
|
||||||
| NC score | 0.998673 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P80404, Q16260, Q8N5W2, Q96BG2, Q99800 | Gene names | ABAT, GABAT | |||
|
Domain Architecture |

|
|||||
| Description | 4-aminobutyrate aminotransferase, mitochondrial precursor (EC 2.6.1.19) ((S)-3-amino-2-methylpropionate transaminase) (EC 2.6.1.22) (Gamma-amino-N-butyrate transaminase) (GABA transaminase) (GABA aminotransferase) (GABA-AT) (GABA-T) (L-AIBAT). | |||||
|
AGT2_HUMAN
|
||||||
| NC score | 0.586737 (rank : 3) | θ value | 4.91457e-14 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BYV1, Q53FB4, Q53FY7, Q53G03, Q5W7Q1 | Gene names | AGXT2, AGT2 | |||
|
Domain Architecture |

|
|||||
| Description | Alanine--glyoxylate aminotransferase 2, mitochondrial precursor (EC 2.6.1.44) ((R)-3-amino-2-methylpropionate--pyruvate transaminase) (EC 2.6.1.40) (AGT 2) (Beta-alanine-pyruvate aminotransferase) (Beta- ALAAT II) (D-AIBAT). | |||||
|
OAT_MOUSE
|
||||||
| NC score | 0.524575 (rank : 4) | θ value | 1.58096e-12 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P29758 | Gene names | Oat | |||
|
Domain Architecture |

|
|||||
| Description | Ornithine aminotransferase, mitochondrial precursor (EC 2.6.1.13) (Ornithine--oxo-acid aminotransferase). | |||||
|
OAT_HUMAN
|
||||||
| NC score | 0.520235 (rank : 5) | θ value | 6.00763e-12 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P04181, Q16068, Q16069, Q9UD03 | Gene names | OAT | |||
|
Domain Architecture |

|
|||||
| Description | Ornithine aminotransferase, mitochondrial precursor (EC 2.6.1.13) (Ornithine--oxo-acid aminotransferase) [Contains: Ornithine aminotransferase, hepatic form; Ornithine aminotransferase, renal form]. | |||||
|
CDC23_MOUSE
|
||||||
| NC score | 0.035067 (rank : 6) | θ value | 3.0926 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BGZ4, Q8C0F1 | Gene names | Cdc23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
CDC23_HUMAN
|
||||||
| NC score | 0.033571 (rank : 7) | θ value | 4.03905 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UJX2, O75433, Q9BS73 | Gene names | CDC23 | |||
|
Domain Architecture |

|
|||||
| Description | Cell division cycle protein 23 homolog (Anaphase-promoting complex subunit 8) (APC8) (Cyclosome subunit 8). | |||||
|
UB2Q2_HUMAN
|
||||||
| NC score | 0.022787 (rank : 8) | θ value | 5.27518 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8WVN8, Q8N4G6, Q96J08 | Gene names | UBE2Q2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-conjugating enzyme E2 Q2 (EC 6.3.2.19) (Ubiquitin-protein ligase Q2) (Ubiquitin carrier protein Q2). | |||||
|
PI52B_HUMAN
|
||||||
| NC score | 0.022573 (rank : 9) | θ value | 8.99809 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78356, Q5U0E8, Q8TBP2 | Gene names | PIP5K2B | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidylinositol-4-phosphate 5-kinase type-2 beta (EC 2.7.1.68) (Phosphatidylinositol-4-phosphate 5-kinase type II beta) (1- phosphatidylinositol-4-phosphate 5-kinase 2-beta) (PtdIns(4)P-5-kinase isoform 2-beta) (PIP5KII-beta) (Diphosphoinositide kinase 2-beta). | |||||
|
TM1L1_HUMAN
|
||||||
| NC score | 0.011032 (rank : 10) | θ value | 8.99809 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75674 | Gene names | TOM1L1, SRCASM | |||
|
Domain Architecture |

|
|||||
| Description | TOM1-like 1 protein (Target of myb-like 1 protein) (Src-activating and signaling molecule protein). | |||||
|
ZN292_HUMAN
|
||||||
| NC score | 0.009939 (rank : 11) | θ value | 6.88961 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60281, Q9H8G3, Q9H8J4 | Gene names | ZNF292, KIAA0530 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 292. | |||||
|
ASB15_MOUSE
|
||||||
| NC score | 0.004333 (rank : 12) | θ value | 8.99809 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHS6 | Gene names | Asb15 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat and SOCS box protein 15 (ASB-15). | |||||