Please be patient as the page loads
|
FEN1_HUMAN
|
||||||
| SwissProt Accessions | P39748 | Gene names | FEN1 | |||
|
Domain Architecture |
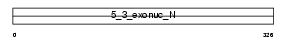
|
|||||
| Description | Flap endonuclease 1 (EC 3.1.-.-) (Flap structure-specific endonuclease 1) (FEN-1) (Maturation factor 1) (MF1) (hFEN-1) (DNase IV). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FEN1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P39748 | Gene names | FEN1 | |||
|
Domain Architecture |
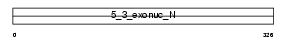
|
|||||
| Description | Flap endonuclease 1 (EC 3.1.-.-) (Flap structure-specific endonuclease 1) (FEN-1) (Maturation factor 1) (MF1) (hFEN-1) (DNase IV). | |||||
|
FEN1_MOUSE
|
||||||
| θ value | 3.82926e-184 (rank : 2) | NC score | 0.998090 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39749 | Gene names | Fen1, Fen-1 | |||
|
Domain Architecture |
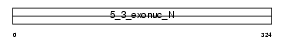
|
|||||
| Description | Flap endonuclease 1 (EC 3.1.-.-) (Flap structure-specific endonuclease 1) (FEN-1). | |||||
|
EXO1_HUMAN
|
||||||
| θ value | 8.65492e-19 (rank : 3) | NC score | 0.588467 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQ84, O60545, O75214, O75466, Q5T396, Q96IJ1, Q9UG38, Q9UNW0 | Gene names | EXO1, EXOI, HEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (hExo1) (Exonuclease I) (hExoI). | |||||
|
EXO1_MOUSE
|
||||||
| θ value | 2.5182e-18 (rank : 4) | NC score | 0.589826 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QZ11, Q3TLM4, Q923A5 | Gene names | Exo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (mExo1) (Exonuclease I). | |||||
|
ERCC5_MOUSE
|
||||||
| θ value | 1.42992e-13 (rank : 5) | NC score | 0.486712 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
ERCC5_HUMAN
|
||||||
| θ value | 9.26847e-13 (rank : 6) | NC score | 0.491835 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 1.38821 (rank : 7) | NC score | 0.015615 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
LRC45_HUMAN
|
||||||
| θ value | 1.81305 (rank : 8) | NC score | 0.024147 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
OLR1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 9) | NC score | 0.026439 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQ09 | Gene names | Olr1, Lox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidized low-density lipoprotein receptor 1 (Ox-LDL receptor 1) (Lectin-type oxidized LDL receptor 1) (Lectin-like oxidized LDL receptor 1) (Lectin-like oxLDL receptor 1) (LOX-1) [Contains: Oxidized low-density lipoprotein receptor 1, soluble form]. | |||||
|
SPAG5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 10) | NC score | 0.017193 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
NIM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 11) | NC score | -0.000093 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IY84 | Gene names | NIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase NIM1 (EC 2.7.11.1). | |||||
|
NUCB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 12) | NC score | 0.019227 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
FEN1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P39748 | Gene names | FEN1 | |||
|
Domain Architecture |
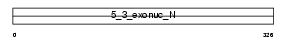
|
|||||
| Description | Flap endonuclease 1 (EC 3.1.-.-) (Flap structure-specific endonuclease 1) (FEN-1) (Maturation factor 1) (MF1) (hFEN-1) (DNase IV). | |||||
|
FEN1_MOUSE
|
||||||
| NC score | 0.998090 (rank : 2) | θ value | 3.82926e-184 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P39749 | Gene names | Fen1, Fen-1 | |||
|
Domain Architecture |
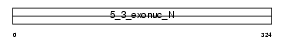
|
|||||
| Description | Flap endonuclease 1 (EC 3.1.-.-) (Flap structure-specific endonuclease 1) (FEN-1). | |||||
|
EXO1_MOUSE
|
||||||
| NC score | 0.589826 (rank : 3) | θ value | 2.5182e-18 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QZ11, Q3TLM4, Q923A5 | Gene names | Exo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (mExo1) (Exonuclease I). | |||||
|
EXO1_HUMAN
|
||||||
| NC score | 0.588467 (rank : 4) | θ value | 8.65492e-19 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9UQ84, O60545, O75214, O75466, Q5T396, Q96IJ1, Q9UG38, Q9UNW0 | Gene names | EXO1, EXOI, HEX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Exonuclease 1 (EC 3.1.-.-) (hExo1) (Exonuclease I) (hExoI). | |||||
|
ERCC5_HUMAN
|
||||||
| NC score | 0.491835 (rank : 5) | θ value | 9.26847e-13 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
ERCC5_MOUSE
|
||||||
| NC score | 0.486712 (rank : 6) | θ value | 1.42992e-13 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35689, Q61528, Q64248 | Gene names | Ercc5, Ercc-5, Xpg | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells homolog (Xeroderma pigmentosum group G-complementing protein homolog) (DNA excision repair protein ERCC-5). | |||||
|
OLR1_MOUSE
|
||||||
| NC score | 0.026439 (rank : 7) | θ value | 2.36792 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 596 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9EQ09 | Gene names | Olr1, Lox1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidized low-density lipoprotein receptor 1 (Ox-LDL receptor 1) (Lectin-type oxidized LDL receptor 1) (Lectin-like oxidized LDL receptor 1) (Lectin-like oxLDL receptor 1) (LOX-1) [Contains: Oxidized low-density lipoprotein receptor 1, soluble form]. | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.024147 (rank : 8) | θ value | 1.81305 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
NUCB2_MOUSE
|
||||||
| NC score | 0.019227 (rank : 9) | θ value | 8.99809 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
SPAG5_HUMAN
|
||||||
| NC score | 0.017193 (rank : 10) | θ value | 3.0926 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96R06, O95213, Q9BWE8, Q9NT17, Q9UFE6 | Gene names | SPAG5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sperm-associated antigen 5 (Astrin) (Mitotic spindle-associated protein p126) (MAP126) (Deepest). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.015615 (rank : 11) | θ value | 1.38821 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
NIM1_HUMAN
|
||||||
| NC score | -0.000093 (rank : 12) | θ value | 8.99809 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 850 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IY84 | Gene names | NIM1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase NIM1 (EC 2.7.11.1). | |||||