Please be patient as the page loads
|
EXOSX_MOUSE
|
||||||
| SwissProt Accessions | P56960, Q9QYS8, Q9R0B1 | Gene names | Exosc10, Pmscl2 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome component 10 (Polymyositis/scleroderma autoantigen 2 homolog) (Autoantigen PM/Scl 2 homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EXOSX_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.989576 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01780, Q15158 | Gene names | EXOSC10, PMSCL, PMSCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome component 10 (Polymyositis/scleroderma autoantigen 2) (Autoantigen PM/Scl 2) (Polymyositis/scleroderma autoantigen 100 kDa) (PM/Scl-100) (P100 polymyositis-scleroderma overlap syndrome- associated autoantigen). | |||||
|
EXOSX_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P56960, Q9QYS8, Q9R0B1 | Gene names | Exosc10, Pmscl2 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome component 10 (Polymyositis/scleroderma autoantigen 2 homolog) (Autoantigen PM/Scl 2 homolog). | |||||
|
CASP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 3) | NC score | 0.029315 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CASP_MOUSE
|
||||||
| θ value | 0.62314 (rank : 4) | NC score | 0.029059 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 5) | NC score | 0.028448 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
EBP2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 6) | NC score | 0.046738 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D903, Q3TKB5 | Gene names | Ebna1bp2, Ebp2 | |||
|
Domain Architecture |
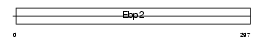
|
|||||
| Description | Probable rRNA-processing protein EBP2. | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 7) | NC score | 0.032273 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
ATF6B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 8) | NC score | 0.046736 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35451 | Gene names | Crebl1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp). | |||||
|
NOTC1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 9) | NC score | 0.008093 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 10) | NC score | 0.013727 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
ESCO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 11) | NC score | 0.023287 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
FAK2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.007943 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14289, Q13475, Q14290, Q16709 | Gene names | PTK2B, FAK2, PYK2, RAFTK | |||
|
Domain Architecture |

|
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
FAK2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.007909 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QVP9 | Gene names | Ptk2b, Fak2, Pyk2, Raftk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
LAMA3_MOUSE
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.011114 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
SDCG8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 15) | NC score | 0.018684 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
BMP6_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.008736 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22004 | Gene names | BMP6, VGR | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 6 precursor (BMP-6). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 17) | NC score | 0.014873 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
PO2F1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 18) | NC score | 0.012770 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P25425, Q61994, Q63891, Q6Y681, Q7TSD0, Q8BT04, Q8K570, Q99JH0, Q9WTZ4, Q9WTZ5, Q9WTZ6 | Gene names | Pou2f1, Oct-1, Otf-1, Otf1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
CALR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.014113 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27797 | Gene names | CALR, CRTC | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) (grp60). | |||||
|
PRD15_HUMAN
|
||||||
| θ value | 5.27518 (rank : 20) | NC score | 0.001225 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57071, Q8N0X3, Q8NEX0, Q9NQV3 | Gene names | PRDM15, C21orf83, ZNF298 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 15 (PR domain-containing protein 15) (Zinc finger protein 298). | |||||
|
RECQ1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 21) | NC score | 0.012530 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 22) | NC score | 0.018882 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 5.27518 (rank : 23) | NC score | 0.018957 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SH24A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 24) | NC score | 0.016876 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D7V1 | Gene names | Sh2d4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A. | |||||
|
NANO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 25) | NC score | 0.019835 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WY41 | Gene names | NANOS1, NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nanos homolog 1 (NOS-1) (EC_Rep1a). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 6.88961 (rank : 26) | NC score | 0.009975 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 6.88961 (rank : 27) | NC score | 0.013288 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
PO2F1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 28) | NC score | 0.010772 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14859, Q5TBT7, Q6PK46, Q8NEU9, Q9BPV1 | Gene names | POU2F1, OCT1, OTF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 29) | NC score | 0.013823 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 30) | NC score | 0.009832 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
MYO5A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 31) | NC score | 0.008539 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
NONO_HUMAN
|
||||||
| θ value | 8.99809 (rank : 32) | NC score | 0.021583 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15233, O00201, P30807, Q12786, Q9BQC5 | Gene names | NONO, NRB54 | |||
|
Domain Architecture |
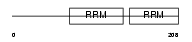
|
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein) (54 kDa nuclear RNA- and DNA-binding protein) (p54(nrb)) (p54nrb) (55 kDa nuclear protein) (NMT55) (DNA-binding p52/p100 complex, 52 kDa subunit). | |||||
|
NONO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 33) | NC score | 0.021530 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99K48, Q63887, Q9CYQ4, Q9DBP2 | Gene names | Nono | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein). | |||||
|
PGCA_HUMAN
|
||||||
| θ value | 8.99809 (rank : 34) | NC score | 0.005053 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 35) | NC score | 0.012477 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
TNR1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 36) | NC score | 0.011145 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19438 | Gene names | TNFRSF1A, TNFAR, TNFR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1A precursor (p60) (TNF-R1) (TNF-RI) (TNFR-I) (p55) (CD120a antigen) [Contains: Tumor necrosis factor receptor superfamily member 1A, membrane form; Tumor necrosis factor-binding protein 1 (TBPI)]. | |||||
|
EXOSX_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P56960, Q9QYS8, Q9R0B1 | Gene names | Exosc10, Pmscl2 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome component 10 (Polymyositis/scleroderma autoantigen 2 homolog) (Autoantigen PM/Scl 2 homolog). | |||||
|
EXOSX_HUMAN
|
||||||
| NC score | 0.989576 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q01780, Q15158 | Gene names | EXOSC10, PMSCL, PMSCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Exosome component 10 (Polymyositis/scleroderma autoantigen 2) (Autoantigen PM/Scl 2) (Polymyositis/scleroderma autoantigen 100 kDa) (PM/Scl-100) (P100 polymyositis-scleroderma overlap syndrome- associated autoantigen). | |||||
|
EBP2_MOUSE
|
||||||
| NC score | 0.046738 (rank : 3) | θ value | 0.813845 (rank : 6) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D903, Q3TKB5 | Gene names | Ebna1bp2, Ebp2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable rRNA-processing protein EBP2. | |||||
|
ATF6B_MOUSE
|
||||||
| NC score | 0.046736 (rank : 4) | θ value | 1.81305 (rank : 8) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35451 | Gene names | Crebl1 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.032273 (rank : 5) | θ value | 1.38821 (rank : 7) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.029315 (rank : 6) | θ value | 0.62314 (rank : 3) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.029059 (rank : 7) | θ value | 0.62314 (rank : 4) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.028448 (rank : 8) | θ value | 0.62314 (rank : 5) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
ESCO1_HUMAN
|
||||||
| NC score | 0.023287 (rank : 9) | θ value | 3.0926 (rank : 11) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q5FWF5, Q69YG4, Q69YS3, Q6IMD7, Q8N3Z5, Q8NBG2, Q96PX7 | Gene names | ESCO1, EFO1, KIAA1911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | N-acetyltransferase ESCO1 (EC 2.3.1.-) (Establishment of cohesion 1 homolog 1) (ECO1 homolog 1) (ESO1 homolog 1) (Establishment factor- like protein 1) (EFO1p) (hEFO1) (CTF7 homolog 1). | |||||
|
NONO_HUMAN
|
||||||
| NC score | 0.021583 (rank : 10) | θ value | 8.99809 (rank : 32) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q15233, O00201, P30807, Q12786, Q9BQC5 | Gene names | NONO, NRB54 | |||
|
Domain Architecture |

|
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein) (54 kDa nuclear RNA- and DNA-binding protein) (p54(nrb)) (p54nrb) (55 kDa nuclear protein) (NMT55) (DNA-binding p52/p100 complex, 52 kDa subunit). | |||||
|
NONO_MOUSE
|
||||||
| NC score | 0.021530 (rank : 11) | θ value | 8.99809 (rank : 33) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q99K48, Q63887, Q9CYQ4, Q9DBP2 | Gene names | Nono | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Non-POU domain-containing octamer-binding protein (NonO protein). | |||||
|
NANO1_HUMAN
|
||||||
| NC score | 0.019835 (rank : 12) | θ value | 6.88961 (rank : 25) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8WY41 | Gene names | NANOS1, NOS1 | |||
|
Domain Architecture |

|
|||||
| Description | Nanos homolog 1 (NOS-1) (EC_Rep1a). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.018957 (rank : 13) | θ value | 5.27518 (rank : 23) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.018882 (rank : 14) | θ value | 5.27518 (rank : 22) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
SDCG8_MOUSE
|
||||||
| NC score | 0.018684 (rank : 15) | θ value | 3.0926 (rank : 15) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q80UF4 | Gene names | Sdccag8, Cccap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (mCCCAP). | |||||
|
SH24A_MOUSE
|
||||||
| NC score | 0.016876 (rank : 16) | θ value | 5.27518 (rank : 24) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9D7V1 | Gene names | Sh2d4a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 4A. | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.014873 (rank : 17) | θ value | 4.03905 (rank : 17) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
CALR_HUMAN
|
||||||
| NC score | 0.014113 (rank : 18) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P27797 | Gene names | CALR, CRTC | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) (grp60). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.013823 (rank : 19) | θ value | 6.88961 (rank : 29) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.013727 (rank : 20) | θ value | 3.0926 (rank : 10) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.013288 (rank : 21) | θ value | 6.88961 (rank : 27) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
PO2F1_MOUSE
|
||||||
| NC score | 0.012770 (rank : 22) | θ value | 4.03905 (rank : 18) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P25425, Q61994, Q63891, Q6Y681, Q7TSD0, Q8BT04, Q8K570, Q99JH0, Q9WTZ4, Q9WTZ5, Q9WTZ6 | Gene names | Pou2f1, Oct-1, Otf-1, Otf1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
RECQ1_MOUSE
|
||||||
| NC score | 0.012530 (rank : 23) | θ value | 5.27518 (rank : 21) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z129, Q9Z128 | Gene names | Recql, Recql1 | |||
|
Domain Architecture |

|
|||||
| Description | ATP-dependent DNA helicase Q1 (EC 3.6.1.-) (DNA-dependent ATPase Q1). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.012477 (rank : 24) | θ value | 8.99809 (rank : 35) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
TNR1A_HUMAN
|
||||||
| NC score | 0.011145 (rank : 25) | θ value | 8.99809 (rank : 36) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P19438 | Gene names | TNFRSF1A, TNFAR, TNFR1 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor necrosis factor receptor superfamily member 1A precursor (p60) (TNF-R1) (TNF-RI) (TNFR-I) (p55) (CD120a antigen) [Contains: Tumor necrosis factor receptor superfamily member 1A, membrane form; Tumor necrosis factor-binding protein 1 (TBPI)]. | |||||
|
LAMA3_MOUSE
|
||||||
| NC score | 0.011114 (rank : 26) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q61789, O08751, Q61788, Q61966, Q9JHQ7 | Gene names | Lama3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-3 chain precursor (Nicein subunit alpha). | |||||
|
PO2F1_HUMAN
|
||||||
| NC score | 0.010772 (rank : 27) | θ value | 6.88961 (rank : 28) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P14859, Q5TBT7, Q6PK46, Q8NEU9, Q9BPV1 | Gene names | POU2F1, OCT1, OTF1 | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 2, transcription factor 1 (Octamer-binding transcription factor 1) (Oct-1) (OTF-1) (NF-A1). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.009975 (rank : 28) | θ value | 6.88961 (rank : 26) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.009832 (rank : 29) | θ value | 8.99809 (rank : 30) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
BMP6_HUMAN
|
||||||
| NC score | 0.008736 (rank : 30) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P22004 | Gene names | BMP6, VGR | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 6 precursor (BMP-6). | |||||
|
MYO5A_MOUSE
|
||||||
| NC score | 0.008539 (rank : 31) | θ value | 8.99809 (rank : 31) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
NOTC1_MOUSE
|
||||||
| NC score | 0.008093 (rank : 32) | θ value | 2.36792 (rank : 9) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q01705, Q06007, Q61905, Q99JC2, Q9QW58, Q9R0X7 | Gene names | Notch1, Motch | |||
|
Domain Architecture |

|
|||||
| Description | Neurogenic locus notch homolog protein 1 precursor (Notch 1) (Motch A) (mT14) (p300) [Contains: Notch 1 extracellular truncation; Notch 1 intracellular domain]. | |||||
|
FAK2_HUMAN
|
||||||
| NC score | 0.007943 (rank : 33) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 847 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q14289, Q13475, Q14290, Q16709 | Gene names | PTK2B, FAK2, PYK2, RAFTK | |||
|
Domain Architecture |

|
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
FAK2_MOUSE
|
||||||
| NC score | 0.007909 (rank : 34) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9QVP9 | Gene names | Ptk2b, Fak2, Pyk2, Raftk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein tyrosine kinase 2 beta (EC 2.7.10.2) (Focal adhesion kinase 2) (FADK 2) (Proline-rich tyrosine kinase 2) (Cell adhesion kinase beta) (CAK beta) (Calcium-dependent tyrosine kinase) (CADTK) (Related adhesion focal tyrosine kinase) (RAFTK). | |||||
|
PGCA_HUMAN
|
||||||
| NC score | 0.005053 (rank : 35) | θ value | 8.99809 (rank : 34) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 821 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P16112, Q13650, Q9UCP4, Q9UCP5, Q9UDE0 | Gene names | AGC1, CSPG1 | |||
|
Domain Architecture |

|
|||||
| Description | Aggrecan core protein precursor (Cartilage-specific proteoglycan core protein) (CSPCP) (Chondroitin sulfate proteoglycan core protein 1) [Contains: Aggrecan core protein 2]. | |||||
|
PRD15_HUMAN
|
||||||
| NC score | 0.001225 (rank : 36) | θ value | 5.27518 (rank : 20) | |||
| Query Neighborhood Hits | 36 | Target Neighborhood Hits | 805 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P57071, Q8N0X3, Q8NEX0, Q9NQV3 | Gene names | PRDM15, C21orf83, ZNF298 | |||
|
Domain Architecture |

|
|||||
| Description | PR domain zinc finger protein 15 (PR domain-containing protein 15) (Zinc finger protein 298). | |||||