Please be patient as the page loads
|
ESTD_HUMAN
|
||||||
| SwissProt Accessions | P10768, Q5TBU8, Q5TBV0, Q5TBV2, Q9BVJ2 | Gene names | ESD | |||
|
Domain Architecture |

|
|||||
| Description | S-formylglutathione hydrolase (EC 3.1.2.12) (FGH) (Esterase D). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ESTD_HUMAN
|
||||||
| θ value | 4.85525e-171 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10768, Q5TBU8, Q5TBV0, Q5TBV2, Q9BVJ2 | Gene names | ESD | |||
|
Domain Architecture |

|
|||||
| Description | S-formylglutathione hydrolase (EC 3.1.2.12) (FGH) (Esterase D). | |||||
|
ESTD_MOUSE
|
||||||
| θ value | 6.1562e-158 (rank : 2) | NC score | 0.994873 (rank : 2) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0P3, Q80ZX4, Q9CWI4 | Gene names | Esd, Es10, Sid478 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | S-formylglutathione hydrolase (EC 3.1.2.12) (FGH) (Esterase D) (Esterase 10) (Sid 478). | |||||
|
DPP10_HUMAN
|
||||||
| θ value | 0.365318 (rank : 3) | NC score | 0.068332 (rank : 4) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N608, Q6TTV4, Q86YR9, Q9P236 | Gene names | DPP10, DPRP3, KIAA1492 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive dipeptidyl peptidase 10 (Dipeptidyl peptidase X) (Dipeptidyl peptidase-like protein 2) (DPL2) (Dipeptidyl peptidase IV-related protein 3) (DPRP-3). | |||||
|
DPP10_MOUSE
|
||||||
| θ value | 0.47712 (rank : 4) | NC score | 0.069572 (rank : 3) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NXK7, Q6P554, Q8K1H0 | Gene names | Dpp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive dipeptidyl peptidase 10 (Dipeptidyl peptidase X). | |||||
|
PTN1_MOUSE
|
||||||
| θ value | 0.47712 (rank : 5) | NC score | 0.018731 (rank : 8) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35821, Q60840, Q62131, Q64498, Q99JS1 | Gene names | Ptpn1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B) (Protein-protein-tyrosine phosphatase HA2) (PTP-HA2). | |||||
|
SEPR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 6) | NC score | 0.062598 (rank : 5) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12884, O00199, Q86Z29, Q99998, Q9UID4 | Gene names | FAP | |||
|
Domain Architecture |
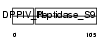
|
|||||
| Description | Seprase (EC 3.4.21.-) (Fibroblast activation protein alpha) (Integral membrane serine protease) (170 kDa melanoma membrane-bound gelatinase). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 7) | NC score | 0.028632 (rank : 7) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
CSPG5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 8) | NC score | 0.015810 (rank : 9) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95196, Q71M39, Q71M40 | Gene names | CSPG5, CALEB, NGC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||
|
SEPR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 9) | NC score | 0.054506 (rank : 6) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97321 | Gene names | Fap | |||
|
Domain Architecture |

|
|||||
| Description | Seprase (EC 3.4.21.-) (Fibroblast activation protein alpha) (Integral membrane serine protease). | |||||
|
ESTD_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.85525e-171 (rank : 1) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P10768, Q5TBU8, Q5TBV0, Q5TBV2, Q9BVJ2 | Gene names | ESD | |||
|
Domain Architecture |

|
|||||
| Description | S-formylglutathione hydrolase (EC 3.1.2.12) (FGH) (Esterase D). | |||||
|
ESTD_MOUSE
|
||||||
| NC score | 0.994873 (rank : 2) | θ value | 6.1562e-158 (rank : 2) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9R0P3, Q80ZX4, Q9CWI4 | Gene names | Esd, Es10, Sid478 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | S-formylglutathione hydrolase (EC 3.1.2.12) (FGH) (Esterase D) (Esterase 10) (Sid 478). | |||||
|
DPP10_MOUSE
|
||||||
| NC score | 0.069572 (rank : 3) | θ value | 0.47712 (rank : 4) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6NXK7, Q6P554, Q8K1H0 | Gene names | Dpp10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive dipeptidyl peptidase 10 (Dipeptidyl peptidase X). | |||||
|
DPP10_HUMAN
|
||||||
| NC score | 0.068332 (rank : 4) | θ value | 0.365318 (rank : 3) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8N608, Q6TTV4, Q86YR9, Q9P236 | Gene names | DPP10, DPRP3, KIAA1492 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inactive dipeptidyl peptidase 10 (Dipeptidyl peptidase X) (Dipeptidyl peptidase-like protein 2) (DPL2) (Dipeptidyl peptidase IV-related protein 3) (DPRP-3). | |||||
|
SEPR_HUMAN
|
||||||
| NC score | 0.062598 (rank : 5) | θ value | 4.03905 (rank : 6) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q12884, O00199, Q86Z29, Q99998, Q9UID4 | Gene names | FAP | |||
|
Domain Architecture |

|
|||||
| Description | Seprase (EC 3.4.21.-) (Fibroblast activation protein alpha) (Integral membrane serine protease) (170 kDa melanoma membrane-bound gelatinase). | |||||
|
SEPR_MOUSE
|
||||||
| NC score | 0.054506 (rank : 6) | θ value | θ > 10 (rank : 9) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97321 | Gene names | Fap | |||
|
Domain Architecture |

|
|||||
| Description | Seprase (EC 3.4.21.-) (Fibroblast activation protein alpha) (Integral membrane serine protease). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.028632 (rank : 7) | θ value | 5.27518 (rank : 7) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
PTN1_MOUSE
|
||||||
| NC score | 0.018731 (rank : 8) | θ value | 0.47712 (rank : 5) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P35821, Q60840, Q62131, Q64498, Q99JS1 | Gene names | Ptpn1 | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 1 (EC 3.1.3.48) (Protein-tyrosine phosphatase 1B) (PTP-1B) (Protein-protein-tyrosine phosphatase HA2) (PTP-HA2). | |||||
|
CSPG5_HUMAN
|
||||||
| NC score | 0.015810 (rank : 9) | θ value | 8.99809 (rank : 8) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O95196, Q71M39, Q71M40 | Gene names | CSPG5, CALEB, NGC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chondroitin sulfate proteoglycan 5 precursor (Neuroglycan C) (Acidic leucine-rich EGF-like domain-containing brain protein). | |||||