Please be patient as the page loads
|
DSG1_HUMAN
|
||||||
| SwissProt Accessions | Q02413 | Gene names | DSG1 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-1 precursor (Desmosomal glycoprotein 1) (DG1) (DGI) (Pemphigus foliaceus antigen). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DSG1A_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998042 (rank : 4) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q61495, Q8CE03 | Gene names | Dsg1a, Dsg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-1 alpha precursor (Dsg1-alpha) (Desmoglein-1) (Desmosomal glycoprotein I) (DG1) (DGI). | |||||
|
DSG1B_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998114 (rank : 2) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q7TSF1, Q7TQ60 | Gene names | Dsg1b, Dsg5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-1 beta precursor (Dsg1-beta) (Desmoglein-5). | |||||
|
DSG1C_MOUSE
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.998095 (rank : 3) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q7TSF0, Q7TQ61 | Gene names | Dsg1c, Dsg6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-1 gamma precursor (Dsg1-gamma) (Desmoglein-6). | |||||
|
DSG1_HUMAN
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 174 | |
| SwissProt Accessions | Q02413 | Gene names | DSG1 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-1 precursor (Desmosomal glycoprotein 1) (DG1) (DGI) (Pemphigus foliaceus antigen). | |||||
|
DSG4_HUMAN
|
||||||
| θ value | 5.18743e-173 (rank : 5) | NC score | 0.986640 (rank : 8) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q86SJ6, Q6Y9L9, Q8IXV4 | Gene names | DSG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-4 precursor. | |||||
|
DSG4_MOUSE
|
||||||
| θ value | 1.2777e-171 (rank : 6) | NC score | 0.987125 (rank : 5) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q7TMD7 | Gene names | Dsg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-4 precursor. | |||||
|
DSG3_HUMAN
|
||||||
| θ value | 1.56915e-153 (rank : 7) | NC score | 0.986793 (rank : 7) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P32926 | Gene names | DSG3 | |||
|
Domain Architecture |
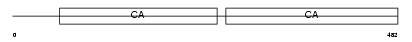
|
|||||
| Description | Desmoglein-3 precursor (130 kDa pemphigus vulgaris antigen) (PVA). | |||||
|
DSG3_MOUSE
|
||||||
| θ value | 1.24331e-150 (rank : 8) | NC score | 0.986925 (rank : 6) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | O35902, Q8CB02, Q8CE48 | Gene names | Dsg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-3 precursor (130 kDa pemphigus vulgaris antigen homolog). | |||||
|
DSG2_HUMAN
|
||||||
| θ value | 4.29305e-135 (rank : 9) | NC score | 0.978678 (rank : 9) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q14126 | Gene names | DSG2 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-2 precursor (HDGC). | |||||
|
DSG2_MOUSE
|
||||||
| θ value | 7.63087e-108 (rank : 10) | NC score | 0.977453 (rank : 10) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | O55111, Q8K069, Q8R517 | Gene names | Dsg2 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-2 precursor. | |||||
|
DSC2_MOUSE
|
||||||
| θ value | 7.45998e-71 (rank : 11) | NC score | 0.954452 (rank : 11) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | P55292, Q64734 | Gene names | Dsc2, Dsc3 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-2 precursor (Epithelial type 2 desmocollin). | |||||
|
DSC2_HUMAN
|
||||||
| θ value | 4.83544e-70 (rank : 12) | NC score | 0.951944 (rank : 13) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q02487 | Gene names | DSC2, DSC3 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-2 precursor (Desmosomal glycoprotein II and III) (Desmocollin-3). | |||||
|
DSC3_HUMAN
|
||||||
| θ value | 3.83135e-67 (rank : 13) | NC score | 0.952350 (rank : 12) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q14574, Q14200, Q9HAZ9 | Gene names | DSC3, DSC4 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-3 precursor (Desmocollin-4) (HT-CP). | |||||
|
DSC3_MOUSE
|
||||||
| θ value | 1.45591e-66 (rank : 14) | NC score | 0.951456 (rank : 14) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55850, O55110, O55122 | Gene names | Dsc3 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-3 precursor. | |||||
|
CADH1_MOUSE
|
||||||
| θ value | 1.45929e-58 (rank : 15) | NC score | 0.949902 (rank : 16) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | P09803, Q61377 | Gene names | Cdh1 | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial-cadherin precursor (E-cadherin) (Uvomorulin) (Cadherin-1) (ARC-1) (CD324 antigen) [Contains: E-Cad/CTF1; E-Cad/CTF2; E- Cad/CTF3]. | |||||
|
CADH4_MOUSE
|
||||||
| θ value | 3.25095e-58 (rank : 16) | NC score | 0.944519 (rank : 28) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P39038 | Gene names | Cdh4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
CADH4_HUMAN
|
||||||
| θ value | 4.24587e-58 (rank : 17) | NC score | 0.944225 (rank : 29) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55283, Q2M208, Q5VZ44, Q9BZ05 | Gene names | CDH4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
CADH1_HUMAN
|
||||||
| θ value | 5.54529e-58 (rank : 18) | NC score | 0.948394 (rank : 19) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | P12830, Q13799, Q14216, Q15855, Q16194, Q4PJ14 | Gene names | CDH1, CDHE, UVO | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial-cadherin precursor (E-cadherin) (Uvomorulin) (Cadherin-1) (CAM 120/80) (CD324 antigen) [Contains: E-Cad/CTF1; E-Cad/CTF2; E- Cad/CTF3]. | |||||
|
DSC1_MOUSE
|
||||||
| θ value | 6.13105e-57 (rank : 19) | NC score | 0.948438 (rank : 18) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55849 | Gene names | Dsc1 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-1 precursor. | |||||
|
CADH2_HUMAN
|
||||||
| θ value | 2.32979e-56 (rank : 20) | NC score | 0.945199 (rank : 26) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P19022, Q14923 | Gene names | CDH2, CDHN, NCAD | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-2 precursor (Neural-cadherin) (N-cadherin) (CDw325 antigen). | |||||
|
DSC1_HUMAN
|
||||||
| θ value | 3.36421e-55 (rank : 21) | NC score | 0.948065 (rank : 20) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q08554 | Gene names | DSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-1 precursor (Desmosomal glycoprotein 2/3) (DG2/DG3). | |||||
|
CADH2_MOUSE
|
||||||
| θ value | 3.71957e-54 (rank : 22) | NC score | 0.944034 (rank : 30) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P15116, Q64260 | Gene names | Cdh2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-2 precursor (Neural-cadherin) (N-cadherin) (CDw325 antigen). | |||||
|
CADH3_MOUSE
|
||||||
| θ value | 3.1488e-53 (rank : 23) | NC score | 0.945965 (rank : 23) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P10287, Q61465 | Gene names | Cdh3, Cdhp | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-3 precursor (Placental-cadherin) (P-cadherin). | |||||
|
CADH3_HUMAN
|
||||||
| θ value | 9.16162e-53 (rank : 24) | NC score | 0.945494 (rank : 25) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P22223 | Gene names | CDH3, CDHP | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-3 precursor (Placental-cadherin) (P-cadherin). | |||||
|
CAD15_HUMAN
|
||||||
| θ value | 1.11993e-50 (rank : 25) | NC score | 0.944661 (rank : 27) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55291 | Gene names | CDH15, CDH14, CDH3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-15 precursor (Muscle-cadherin) (M-cadherin) (Cadherin-14). | |||||
|
CAD15_MOUSE
|
||||||
| θ value | 7.25919e-50 (rank : 26) | NC score | 0.945523 (rank : 24) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | P33146, Q9QYZ7 | Gene names | Cdh15, Cdh14 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-15 precursor (Muscle-cadherin) (M-cadherin) (Cadherin-14). | |||||
|
CAD13_HUMAN
|
||||||
| θ value | 3.60269e-49 (rank : 27) | NC score | 0.947284 (rank : 21) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55290, Q8TBX3 | Gene names | CDH13, CDHH | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-13 precursor (Truncated-cadherin) (T-cadherin) (T-cad) (Heart-cadherin) (H-cadherin) (P105). | |||||
|
CAD13_MOUSE
|
||||||
| θ value | 3.60269e-49 (rank : 28) | NC score | 0.946789 (rank : 22) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9WTR5 | Gene names | Cdh13 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-13 precursor (Truncated-cadherin) (T-cadherin) (T-cad) (Heart-cadherin) (H-cadherin). | |||||
|
CAD26_HUMAN
|
||||||
| θ value | 2.85459e-46 (rank : 29) | NC score | 0.948567 (rank : 17) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q8IXH8, Q8TCH3, Q9BQN4, Q9NRU1 | Gene names | CDH26 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-like protein 26 precursor (Cadherin-like protein VR20). | |||||
|
CAD26_MOUSE
|
||||||
| θ value | 6.58091e-43 (rank : 30) | NC score | 0.949962 (rank : 15) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P59862 | Gene names | Cdh26 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-like protein 26 precursor. | |||||
|
CAD12_HUMAN
|
||||||
| θ value | 5.21438e-40 (rank : 31) | NC score | 0.907613 (rank : 35) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55289 | Gene names | CDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-12 precursor (Brain-cadherin) (BR-cadherin) (N-cadherin 2) (Neural type cadherin 2). | |||||
|
CADH6_HUMAN
|
||||||
| θ value | 3.16345e-37 (rank : 32) | NC score | 0.904821 (rank : 38) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55285, Q9BWS0 | Gene names | CDH6 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-6 precursor (Kidney-cadherin) (K-cadherin). | |||||
|
CAD11_HUMAN
|
||||||
| θ value | 5.96599e-36 (rank : 33) | NC score | 0.904319 (rank : 39) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55287, Q15065, Q15066, Q9UQ93, Q9UQ94 | Gene names | CDH11 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-11 precursor (Osteoblast-cadherin) (OB-cadherin) (OSF-4). | |||||
|
CAD11_MOUSE
|
||||||
| θ value | 7.79185e-36 (rank : 34) | NC score | 0.904166 (rank : 40) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55288 | Gene names | Cdh11, Cad-11 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-11 precursor (Osteoblast-cadherin) (OB-cadherin) (OSF-4). | |||||
|
CAD18_HUMAN
|
||||||
| θ value | 1.73584e-35 (rank : 35) | NC score | 0.907323 (rank : 36) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q13634 | Gene names | CDH18, CDH14 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-18 precursor (Cadherin-14). | |||||
|
CADH6_MOUSE
|
||||||
| θ value | 3.86705e-35 (rank : 36) | NC score | 0.902960 (rank : 41) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P97326, P70393 | Gene names | Cdh6 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-6 precursor (Kidney-cadherin) (K-cadherin). | |||||
|
CAD10_HUMAN
|
||||||
| θ value | 2.50655e-34 (rank : 37) | NC score | 0.902624 (rank : 43) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y6N8, Q9ULB3 | Gene names | CDH10 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-10 precursor (T2-cadherin). | |||||
|
CAD20_HUMAN
|
||||||
| θ value | 4.27553e-34 (rank : 38) | NC score | 0.902750 (rank : 42) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9HBT6 | Gene names | CDH20, CDH7L3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-20 precursor. | |||||
|
CAD17_HUMAN
|
||||||
| θ value | 8.06329e-33 (rank : 39) | NC score | 0.923869 (rank : 31) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q12864, Q15336 | Gene names | CDH17 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-17 precursor (Liver-intestine-cadherin) (LI-cadherin) (Intestinal peptide-associated transporter HPT-1). | |||||
|
CAD17_MOUSE
|
||||||
| θ value | 8.06329e-33 (rank : 40) | NC score | 0.922484 (rank : 32) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9R100 | Gene names | Cdh17 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-17 precursor (Liver-intestine-cadherin) (LI-cadherin) (BILL- cadherin) (P130). | |||||
|
CAD24_HUMAN
|
||||||
| θ value | 3.06405e-32 (rank : 41) | NC score | 0.902512 (rank : 44) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q86UP0, Q86UP1, Q9NT84 | Gene names | CDH24, CDH11L | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-24 precursor. | |||||
|
CADH7_HUMAN
|
||||||
| θ value | 3.06405e-32 (rank : 42) | NC score | 0.905219 (rank : 37) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9ULB5, Q9H157 | Gene names | CDH7, CDH7L1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-7 precursor. | |||||
|
CADH5_HUMAN
|
||||||
| θ value | 1.98606e-31 (rank : 43) | NC score | 0.908323 (rank : 33) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P33151 | Gene names | CDH5 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-5 precursor (Vascular endothelial-cadherin) (VE-cadherin) (7B4 antigen) (CD144 antigen). | |||||
|
CADH5_MOUSE
|
||||||
| θ value | 9.8567e-31 (rank : 44) | NC score | 0.907710 (rank : 34) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55284, O35542 | Gene names | Cdh5 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-5 precursor (Vascular endothelial-cadherin) (VE-cadherin) (CD144 antigen). | |||||
|
CADH9_HUMAN
|
||||||
| θ value | 3.74554e-30 (rank : 45) | NC score | 0.898220 (rank : 47) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9ULB4 | Gene names | CDH9 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-9 precursor. | |||||
|
CAD22_MOUSE
|
||||||
| θ value | 2.42779e-29 (rank : 46) | NC score | 0.900711 (rank : 45) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9WTP5 | Gene names | Cdh22 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-22 precursor (Pituitary and brain cadherin) (PB-cadherin). | |||||
|
CADH8_HUMAN
|
||||||
| θ value | 1.57365e-28 (rank : 47) | NC score | 0.897029 (rank : 49) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55286, Q9ULB2 | Gene names | CDH8 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-8 precursor. | |||||
|
CAD19_HUMAN
|
||||||
| θ value | 2.05525e-28 (rank : 48) | NC score | 0.897919 (rank : 48) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9H159 | Gene names | CDH19, CDH7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-19 precursor. | |||||
|
CADH8_MOUSE
|
||||||
| θ value | 3.50572e-28 (rank : 49) | NC score | 0.896635 (rank : 50) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P97291 | Gene names | Cdh8 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-8 precursor. | |||||
|
CAD22_HUMAN
|
||||||
| θ value | 5.06226e-27 (rank : 50) | NC score | 0.898359 (rank : 46) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9UJ99, O43205 | Gene names | CDH22, C20orf25 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-22 precursor (Pituitary and brain cadherin) (PB-cadherin). | |||||
|
CELR1_HUMAN
|
||||||
| θ value | 2.12685e-25 (rank : 51) | NC score | 0.696100 (rank : 134) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
FATH_HUMAN
|
||||||
| θ value | 4.73814e-25 (rank : 52) | NC score | 0.745642 (rank : 128) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q14517 | Gene names | FAT | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-related tumor suppressor homolog precursor (Protein fat homolog). | |||||
|
CELR1_MOUSE
|
||||||
| θ value | 7.56453e-23 (rank : 53) | NC score | 0.696722 (rank : 133) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | O35161 | Gene names | Celsr1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor. | |||||
|
CAD23_HUMAN
|
||||||
| θ value | 9.87957e-23 (rank : 54) | NC score | 0.838205 (rank : 54) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9H251, Q6UWW1, Q9H4K9 | Gene names | CDH23, KIAA1774 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
CAD23_MOUSE
|
||||||
| θ value | 9.87957e-23 (rank : 55) | NC score | 0.838060 (rank : 55) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q99PF4, Q99NH1, Q9D4N9 | Gene names | Cdh23 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
PCDAB_HUMAN
|
||||||
| θ value | 1.09232e-21 (rank : 56) | NC score | 0.771964 (rank : 90) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9Y5I1, O75279 | Gene names | PCDHA11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 11 precursor (PCDH-alpha11). | |||||
|
CELR3_HUMAN
|
||||||
| θ value | 1.2077e-20 (rank : 57) | NC score | 0.697970 (rank : 132) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
PCDG2_HUMAN
|
||||||
| θ value | 1.5773e-20 (rank : 58) | NC score | 0.770776 (rank : 95) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5H1, Q9Y5D5 | Gene names | PCDHGA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A2 precursor (PCDH-gamma-A2). | |||||
|
CELR3_MOUSE
|
||||||
| θ value | 2.06002e-20 (rank : 59) | NC score | 0.699108 (rank : 131) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
FAT2_HUMAN
|
||||||
| θ value | 2.06002e-20 (rank : 60) | NC score | 0.792674 (rank : 66) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
PCDA1_HUMAN
|
||||||
| θ value | 2.69047e-20 (rank : 61) | NC score | 0.768596 (rank : 101) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9Y5I3, O75288, Q9NRT7 | Gene names | PCDHA1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 1 precursor (PCDH-alpha1). | |||||
|
PCDA5_HUMAN
|
||||||
| θ value | 2.69047e-20 (rank : 62) | NC score | 0.767219 (rank : 108) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9Y5H7, O75284, Q8N4R3 | Gene names | PCDHA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 5 precursor (PCDH-alpha5). | |||||
|
CELR2_HUMAN
|
||||||
| θ value | 3.51386e-20 (rank : 63) | NC score | 0.694571 (rank : 135) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
PCDBC_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 64) | NC score | 0.778417 (rank : 74) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5F1 | Gene names | PCDHB12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 12 precursor (PCDH-beta12). | |||||
|
PCDH1_HUMAN
|
||||||
| θ value | 7.82807e-20 (rank : 65) | NC score | 0.814427 (rank : 59) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q08174 | Gene names | PCDH1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-1 precursor (Protocadherin-42) (PC42) (Cadherin-like protein 1). | |||||
|
PCDB6_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 66) | NC score | 0.777664 (rank : 78) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5E3 | Gene names | PCDHB6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 6 precursor (PCDH-beta6). | |||||
|
PCDGH_HUMAN
|
||||||
| θ value | 1.02238e-19 (rank : 67) | NC score | 0.779776 (rank : 71) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G0, Q9Y5C6 | Gene names | PCDHGB5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B5 precursor (PCDH-gamma-B5). | |||||
|
PCDB5_HUMAN
|
||||||
| θ value | 1.33526e-19 (rank : 68) | NC score | 0.770607 (rank : 97) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q9Y5E4, Q9UFU9 | Gene names | PCDHB5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 5 precursor (PCDH-beta5). | |||||
|
PCDBG_HUMAN
|
||||||
| θ value | 1.33526e-19 (rank : 69) | NC score | 0.777078 (rank : 79) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9NRJ7, Q8IYD5, Q96SE9, Q9HCF1 | Gene names | PCDHB16, KIAA1621, PCDH3X | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 16 precursor (PCDH-beta16) (Protocadherin 3X). | |||||
|
CELR2_MOUSE
|
||||||
| θ value | 1.74391e-19 (rank : 70) | NC score | 0.700921 (rank : 130) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
PC11Y_HUMAN
|
||||||
| θ value | 2.27762e-19 (rank : 71) | NC score | 0.803482 (rank : 64) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q9BZA8, Q70LR6, Q70LR8, Q70LS0, Q70LS1, Q70LS2, Q70LS3, Q70LS4, Q70LS5, Q8WY34, Q9BZA9, Q9H4E1 | Gene names | PCDH11Y, PCDH11, PCDH22, PCDHY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 Y-linked precursor (Protocadherin-11) (Protocadherin- 22) (Protocadherin on the Y chromosome) (PCDH-Y) (Protocadherin prostate cancer) (Protocadherin-PC). | |||||
|
CAD16_HUMAN
|
||||||
| θ value | 3.88503e-19 (rank : 72) | NC score | 0.891196 (rank : 52) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | O75309, Q6UW93 | Gene names | CDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-16 precursor (Kidney-specific cadherin) (Ksp-cadherin). | |||||
|
PCDBB_HUMAN
|
||||||
| θ value | 3.88503e-19 (rank : 73) | NC score | 0.773545 (rank : 86) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5F2 | Gene names | PCDHB11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 11 precursor (PCDH-beta11). | |||||
|
PCDG6_HUMAN
|
||||||
| θ value | 3.88503e-19 (rank : 74) | NC score | 0.767988 (rank : 102) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G7, Q9Y5D1 | Gene names | PCDHGA6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A6 precursor (PCDH-gamma-A6). | |||||
|
PCDGK_HUMAN
|
||||||
| θ value | 6.62687e-19 (rank : 75) | NC score | 0.774310 (rank : 83) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9UN70, O60622, Q08192, Q9Y5C4 | Gene names | PCDHGC3, PCDH2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C3 precursor (PCDH-gamma-C3) (Protocadherin 43) (PC-43) (Protocadherin 2). | |||||
|
PCDC2_HUMAN
|
||||||
| θ value | 8.65492e-19 (rank : 76) | NC score | 0.785667 (rank : 67) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9Y5I4, Q9Y5F4 | Gene names | PCDHAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C2 precursor (PCDH-alpha-C2). | |||||
|
PC11X_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 77) | NC score | 0.801854 (rank : 65) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q9BZA7, Q2TJH0, Q2TJH1, Q2TJH3, Q5JVZ0, Q70LR8, Q70LS7, Q70LS8, Q70LS9, Q70LT7, Q70LT8, Q70LT9, Q70LU0, Q70LU1, Q96RV4, Q96RW0, Q9BZA6, Q9H4E0, Q9P2M0, Q9P2X5 | Gene names | PCDH11X, PCDH11, PCDHX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 X-linked precursor (Protocadherin-11) (Protocadherin on the X chromosome) (PCDH-X) (Protocadherin-S). | |||||
|
PCDA2_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 78) | NC score | 0.771290 (rank : 93) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9Y5H9, O75287, Q9BTV3 | Gene names | PCDHA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 2 precursor (PCDH-alpha2). | |||||
|
PCDGC_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 79) | NC score | 0.767694 (rank : 104) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | O60330, O15100, Q6UW70, Q9Y5D7 | Gene names | PCDHGA12, CDH21, FIB3, KIAA0588 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A12 precursor (PCDH-gamma-A12) (Cadherin-21) (Fibroblast cadherin 3). | |||||
|
PCDGF_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 80) | NC score | 0.771569 (rank : 92) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G1, Q9Y5C7 | Gene names | PCDHGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B3 precursor (PCDH-gamma-B3). | |||||
|
PCDBF_HUMAN
|
||||||
| θ value | 1.92812e-18 (rank : 81) | NC score | 0.774133 (rank : 84) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q9Y5E8, Q8IUX5 | Gene names | PCDHB15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 15 precursor (PCDH-beta15). | |||||
|
PCDB9_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 82) | NC score | 0.774476 (rank : 82) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5E1, Q9NRJ8 | Gene names | PCDHB9, PCDH3H | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 9 precursor (PCDH-beta9) (Protocadherin 3H). | |||||
|
PCDGA_HUMAN
|
||||||
| θ value | 3.28887e-18 (rank : 83) | NC score | 0.764292 (rank : 116) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5H3, Q9Y5E0 | Gene names | PCDHGA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A10 precursor (PCDH-gamma-A10). | |||||
|
PCDGB_HUMAN
|
||||||
| θ value | 4.29542e-18 (rank : 84) | NC score | 0.771080 (rank : 94) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5H2, Q9Y5D8, Q9Y5D9 | Gene names | PCDHGA11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A11 precursor (PCDH-gamma-A11). | |||||
|
PCDGD_HUMAN
|
||||||
| θ value | 4.29542e-18 (rank : 85) | NC score | 0.776740 (rank : 80) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G3, Q9Y5C8 | Gene names | PCDHGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B1 precursor (PCDH-gamma-B1). | |||||
|
PCDGG_HUMAN
|
||||||
| θ value | 4.29542e-18 (rank : 86) | NC score | 0.781978 (rank : 68) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9UN71, O15099, Q9UN64 | Gene names | PCDHGB4, CDH20, FIB2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B4 precursor (PCDH-gamma-B4) (Cadherin-20) (Fibroblast cadherin 2). | |||||
|
PCDG1_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 87) | NC score | 0.767657 (rank : 105) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5H4, Q9Y5D6 | Gene names | PCDHGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A1 precursor (PCDH-gamma-A1). | |||||
|
PCDBE_HUMAN
|
||||||
| θ value | 9.56915e-18 (rank : 88) | NC score | 0.771810 (rank : 91) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q9Y5E9 | Gene names | PCDHB14 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 14 precursor (PCDH-beta14). | |||||
|
PCD16_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 89) | NC score | 0.821439 (rank : 58) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q96JQ0, O15098 | Gene names | DCHS1, CDH19, FIB1, KIAA1773, PCDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-16 precursor (Dachsous 1) (Cadherin-19) (Fibroblast cadherin 1). | |||||
|
PCDA4_MOUSE
|
||||||
| θ value | 1.24977e-17 (rank : 90) | NC score | 0.766641 (rank : 111) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 146 | |
| SwissProt Accessions | O88689, Q3UEX3, Q6PAM9, Q8K487, Q8K489 | Gene names | Pcdha4, Cnr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin alpha 4 precursor (PCDH-alpha4). | |||||
|
PCDH7_HUMAN
|
||||||
| θ value | 1.24977e-17 (rank : 91) | NC score | 0.808551 (rank : 60) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | O60245, O60246, O60247 | Gene names | PCDH7, BHPCDH | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-7 precursor (Brain-heart protocadherin) (BH-Pcdh). | |||||
|
PCDB7_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 92) | NC score | 0.775106 (rank : 81) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5E2 | Gene names | PCDHB7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 7 precursor (PCDH-beta7). | |||||
|
PCDB8_HUMAN
|
||||||
| θ value | 1.63225e-17 (rank : 93) | NC score | 0.772936 (rank : 87) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9UN66 | Gene names | PCDHB8, PCDH3I | |||
|
Domain Architecture |
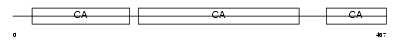
|
|||||
| Description | Protocadherin beta 8 precursor (PCDH-beta8) (Protocadherin 3I). | |||||
|
CAD16_MOUSE
|
||||||
| θ value | 2.13179e-17 (rank : 94) | NC score | 0.893444 (rank : 51) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | O88338, Q9JLZ5 | Gene names | Cdh16 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-16 precursor (Kidney-specific cadherin) (Ksp-cadherin). | |||||
|
PCD12_HUMAN
|
||||||
| θ value | 2.13179e-17 (rank : 95) | NC score | 0.779277 (rank : 72) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q9NPG4, Q6UXB6, Q96KB8, Q9H7Y6, Q9H8E0 | Gene names | PCDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
PCDA3_HUMAN
|
||||||
| θ value | 2.13179e-17 (rank : 96) | NC score | 0.760675 (rank : 121) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9Y5H8, O75286 | Gene names | PCDHA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 3 precursor (PCDH-alpha3). | |||||
|
PCDAD_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 97) | NC score | 0.766502 (rank : 112) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9Y5I0, O75277 | Gene names | PCDHA13 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 13 precursor (PCDH-alpha13). | |||||
|
PCDG9_HUMAN
|
||||||
| θ value | 2.7842e-17 (rank : 98) | NC score | 0.772005 (rank : 89) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G4, Q9Y5C9 | Gene names | PCDHGA9 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A9 precursor (PCDH-gamma-A9). | |||||
|
PCDG7_HUMAN
|
||||||
| θ value | 3.63628e-17 (rank : 99) | NC score | 0.765599 (rank : 115) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G6, Q9Y5D0 | Gene names | PCDHGA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A7 precursor (PCDH-gamma-A7). | |||||
|
PCDB2_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 100) | NC score | 0.767929 (rank : 103) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q9Y5E7 | Gene names | PCDHB2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 2 precursor (PCDH-beta2). | |||||
|
PCDG8_HUMAN
|
||||||
| θ value | 4.74913e-17 (rank : 101) | NC score | 0.769087 (rank : 100) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G5, O15039 | Gene names | PCDHGA8, KIAA0327 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A8 precursor (PCDH-gamma-A8). | |||||
|
PCD20_HUMAN
|
||||||
| θ value | 8.10077e-17 (rank : 102) | NC score | 0.807299 (rank : 61) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q8N6Y1, Q8NDN4, Q9NRT9 | Gene names | PCDH20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-20 precursor (Protocadherin-13). | |||||
|
PCD10_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 103) | NC score | 0.778409 (rank : 75) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9P2E7 | Gene names | PCDH10, KIAA1400 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-10 precursor. | |||||
|
PCDBA_HUMAN
|
||||||
| θ value | 1.058e-16 (rank : 104) | NC score | 0.767311 (rank : 107) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9UN67, Q96T99 | Gene names | PCDHB10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 10 precursor (PCDH-beta10). | |||||
|
PCDA8_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 105) | NC score | 0.761965 (rank : 118) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9Y5H6, O75281 | Gene names | PCDHA8 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 8 precursor (PCDH-alpha8). | |||||
|
PCDAA_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 106) | NC score | 0.753320 (rank : 127) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9Y5I2, O75280, Q9NRU2 | Gene names | PCDHA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 10 precursor (PCDH-alpha10). | |||||
|
PCDH9_HUMAN
|
||||||
| θ value | 1.38178e-16 (rank : 107) | NC score | 0.804897 (rank : 63) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9HC56 | Gene names | PCDH9 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-9 precursor. | |||||
|
PCDG5_HUMAN
|
||||||
| θ value | 1.80466e-16 (rank : 108) | NC score | 0.766132 (rank : 113) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G8, Q9Y5D2 | Gene names | PCDHGA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A5 precursor (PCDH-gamma-A5). | |||||
|
PCDG3_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 109) | NC score | 0.767648 (rank : 106) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5H0, Q9Y5D4 | Gene names | PCDHGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A3 precursor (PCDH-gamma-A3). | |||||
|
PCDLK_HUMAN
|
||||||
| θ value | 4.02038e-16 (rank : 110) | NC score | 0.848945 (rank : 53) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9BYE9, Q9NXP8 | Gene names | PCLKC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin LKC precursor (PC-LKC). | |||||
|
PCD17_HUMAN
|
||||||
| θ value | 5.25075e-16 (rank : 111) | NC score | 0.778697 (rank : 73) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | O14917 | Gene names | PCDH17, PCDH68, PCH68 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-17 precursor (Protocadherin-68). | |||||
|
PCDA6_HUMAN
|
||||||
| θ value | 5.25075e-16 (rank : 112) | NC score | 0.760830 (rank : 120) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9UN73, O75283, Q9NRT8 | Gene names | PCDHA6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 6 precursor (PCDH-alpha6). | |||||
|
PCDB3_HUMAN
|
||||||
| θ value | 5.25075e-16 (rank : 113) | NC score | 0.766847 (rank : 110) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5E6 | Gene names | PCDHB3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 3 precursor (PCDH-beta3). | |||||
|
PCDB1_HUMAN
|
||||||
| θ value | 8.95645e-16 (rank : 114) | NC score | 0.766052 (rank : 114) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5F3 | Gene names | PCDHB1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 1 precursor (PCDH-beta1). | |||||
|
PCD12_MOUSE
|
||||||
| θ value | 1.16975e-15 (rank : 115) | NC score | 0.780238 (rank : 70) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | O55134 | Gene names | Pcdh12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
PCDGE_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 116) | NC score | 0.767074 (rank : 109) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G2, Q9UN65 | Gene names | PCDHGB2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B2 precursor (PCDH-gamma-B2). | |||||
|
PCDGM_HUMAN
|
||||||
| θ value | 1.52774e-15 (rank : 117) | NC score | 0.763012 (rank : 117) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 141 | |
| SwissProt Accessions | Q9Y5F6, Q9Y5C2 | Gene names | PCDHGC5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C5 precursor (PCDH-gamma-C5). | |||||
|
PCDB4_HUMAN
|
||||||
| θ value | 1.99529e-15 (rank : 118) | NC score | 0.773874 (rank : 85) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5E5 | Gene names | PCDHB4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 4 precursor (PCDH-beta4). | |||||
|
PCDBD_HUMAN
|
||||||
| θ value | 1.99529e-15 (rank : 119) | NC score | 0.770610 (rank : 96) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5F0 | Gene names | PCDHB13 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 13 precursor (PCDH-beta13). | |||||
|
PCDG4_HUMAN
|
||||||
| θ value | 1.99529e-15 (rank : 120) | NC score | 0.761583 (rank : 119) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G9, Q9Y5D3 | Gene names | PCDHGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A4 precursor (PCDH-gamma-A4). | |||||
|
PCDA9_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 121) | NC score | 0.757518 (rank : 124) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9Y5H5, O15053 | Gene names | PCDHA9, KIAA0345 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 9 precursor (PCDH-alpha9). | |||||
|
PCDGI_HUMAN
|
||||||
| θ value | 3.40345e-15 (rank : 122) | NC score | 0.769422 (rank : 98) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5F9, Q9Y5C5 | Gene names | PCDHGB6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B6 precursor (PCDH-gamma-B6). | |||||
|
PCD18_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 123) | NC score | 0.781309 (rank : 69) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9HCL0 | Gene names | PCDH18, KIAA1562 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-18 precursor. | |||||
|
PCDA7_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 124) | NC score | 0.753949 (rank : 126) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9UN72, O75282 | Gene names | PCDHA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 7 precursor (PCDH-alpha7). | |||||
|
PCDAC_HUMAN
|
||||||
| θ value | 2.88119e-14 (rank : 125) | NC score | 0.758059 (rank : 123) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9UN75, O75278 | Gene names | PCDHA12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 12 precursor (PCDH-alpha12). | |||||
|
PCDGJ_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 126) | NC score | 0.772175 (rank : 88) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5F8, Q9UN63 | Gene names | PCDHGB7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B7 precursor (PCDH-gamma-B7). | |||||
|
PCD15_HUMAN
|
||||||
| θ value | 1.86753e-13 (rank : 127) | NC score | 0.823137 (rank : 57) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
PCD15_MOUSE
|
||||||
| θ value | 2.43908e-13 (rank : 128) | NC score | 0.825564 (rank : 56) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q99PJ1 | Gene names | Pcdh15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
PCDGL_HUMAN
|
||||||
| θ value | 2.43908e-13 (rank : 129) | NC score | 0.769119 (rank : 99) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5F7, Q9Y5C3 | Gene names | PCDHGC4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C4 precursor (PCDH-gamma-C4). | |||||
|
PCD20_MOUSE
|
||||||
| θ value | 3.18553e-13 (rank : 130) | NC score | 0.804928 (rank : 62) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q8BIZ0, Q8BIV2 | Gene names | Pcdh20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-20 precursor. | |||||
|
PCD19_HUMAN
|
||||||
| θ value | 4.16044e-13 (rank : 131) | NC score | 0.778085 (rank : 77) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q8TAB3, Q5JTG1, Q5JTG2, Q68DT7, Q9P2N3 | Gene names | PCDH19, KIAA1313 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-19 precursor. | |||||
|
PCD19_MOUSE
|
||||||
| θ value | 2.69671e-12 (rank : 132) | NC score | 0.778086 (rank : 76) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q80TF3, Q8BL13 | Gene names | Pcdh19, Kiaa1313 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-19 precursor. | |||||
|
PCDC1_HUMAN
|
||||||
| θ value | 1.74796e-11 (rank : 133) | NC score | 0.758573 (rank : 122) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9H158, Q9Y5F5, Q9Y5I5 | Gene names | PCDHAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C1 precursor (PCDH-alpha-C1). | |||||
|
PCDA4_HUMAN
|
||||||
| θ value | 2.28291e-11 (rank : 134) | NC score | 0.755181 (rank : 125) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9UN74, O75285 | Gene names | PCDHA4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 4 precursor (PCDH-alpha4). | |||||
|
PCDH8_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 135) | NC score | 0.740786 (rank : 129) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | O95206, Q8IYE9, Q96SF1 | Gene names | PCDH8 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-8 precursor (Arcadlin). | |||||
|
CSTN1_MOUSE
|
||||||
| θ value | 1.87187e-05 (rank : 136) | NC score | 0.515880 (rank : 138) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q9EPL2 | Gene names | Clstn1, Cs1, Cstn1 | |||
|
Domain Architecture |
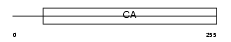
|
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
CSTN1_HUMAN
|
||||||
| θ value | 0.000158464 (rank : 137) | NC score | 0.491045 (rank : 140) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | O94985 | Gene names | CLSTN1, CS1, KIAA0911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
CSTN2_HUMAN
|
||||||
| θ value | 0.000270298 (rank : 138) | NC score | 0.491022 (rank : 141) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | Q9H4D0, Q9BSS0 | Gene names | CLSTN2, CS2 | |||
|
Domain Architecture |
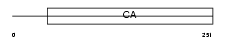
|
|||||
| Description | Calsyntenin-2 precursor. | |||||
|
CSTN2_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 139) | NC score | 0.495183 (rank : 139) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | Q9ER65 | Gene names | Clstn2, Cs2, Cstn2 | |||
|
Domain Architecture |
|
|||||
| Description | Calsyntenin-2 precursor. | |||||
|
CSTN3_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 140) | NC score | 0.426566 (rank : 142) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q9BQT9, O94831 | Gene names | CLSTN3, CS3, KIAA0726 | |||
|
Domain Architecture |
|
|||||
| Description | Calsyntenin-3 precursor. | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 141) | NC score | 0.600883 (rank : 136) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 141 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
MUCDL_HUMAN
|
||||||
| θ value | 0.125558 (rank : 142) | NC score | 0.548875 (rank : 137) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
MAGI1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 143) | NC score | 0.002266 (rank : 162) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
PER1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 144) | NC score | 0.008400 (rank : 152) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 0.47712 (rank : 145) | NC score | 0.006128 (rank : 154) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
SON_MOUSE
|
||||||
| θ value | 1.06291 (rank : 146) | NC score | 0.003898 (rank : 158) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
CBP_HUMAN
|
||||||
| θ value | 1.38821 (rank : 147) | NC score | 0.009357 (rank : 151) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
MARCO_MOUSE
|
||||||
| θ value | 1.81305 (rank : 148) | NC score | -0.000124 (rank : 167) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
PXDC1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 149) | NC score | 0.007429 (rank : 153) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IUK5, Q5QCZ7, Q5QCZ8, Q5QCZ9, Q9HCT9 | Gene names | PLXDC1, TEM3, TEM7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 1 precursor (Tumor endothelial marker 7) (Tumor endothelial marker 3). | |||||
|
ZN452_HUMAN
|
||||||
| θ value | 2.36792 (rank : 150) | NC score | 0.001242 (rank : 163) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6R2W3, Q2NKL9, Q5SRJ3, Q8TCN2, Q96PW3 | Gene names | ZNF452, KIAA1925 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF452. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 151) | NC score | 0.016714 (rank : 150) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CAN2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 152) | NC score | 0.003333 (rank : 160) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17655, Q16738, Q6PJT3, Q8WU26, Q9HBB1 | Gene names | CAPN2, CANPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (Calpain large polypeptide L2). | |||||
|
CSPG2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 153) | NC score | 0.031688 (rank : 149) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
NR4A3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 154) | NC score | -0.000972 (rank : 168) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92570, Q12935, Q14979, Q16420, Q4VXA9, Q9UEK2, Q9UEK3 | Gene names | NR4A3, CHN, CSMF, MINOR, NOR1, TEC | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A3 (Nuclear hormone receptor NOR-1) (Neuron-derived orphan receptor 1) (Mitogen-induced nuclear orphan receptor). | |||||
|
PO121_HUMAN
|
||||||
| θ value | 4.03905 (rank : 155) | NC score | 0.003886 (rank : 159) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
TAF9_MOUSE
|
||||||
| θ value | 4.03905 (rank : 156) | NC score | 0.005814 (rank : 155) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VI33, Q32P09, Q80XS3, Q9CV61 | Gene names | Taf9, Taf2g | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 9 (Transcription initiation factor TFIID 31 kDa subunit) (TAFII-31) (TAFII-32) (TAFII32). | |||||
|
CO4A2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 157) | NC score | -0.001384 (rank : 170) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
SMRC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 158) | NC score | 0.000070 (rank : 166) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
CAN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 159) | NC score | 0.002510 (rank : 161) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08529, O35518, O54843 | Gene names | Capn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (80 kDa M-calpain subunit) (CALP80). | |||||
|
FA29A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 160) | NC score | 0.005624 (rank : 156) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z4H7, Q6IQ10, Q6NZX5, Q8IZQ4, Q96FN0, Q9H950, Q9H998, Q9HCJ8, Q9NXT8 | Gene names | FAM29A, KIAA1574 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM29A. | |||||
|
MINT_HUMAN
|
||||||
| θ value | 6.88961 (rank : 161) | NC score | -0.003314 (rank : 176) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
TS101_HUMAN
|
||||||
| θ value | 6.88961 (rank : 162) | NC score | 0.000414 (rank : 165) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 163) | NC score | -0.010549 (rank : 180) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ATS14_HUMAN
|
||||||
| θ value | 8.99809 (rank : 164) | NC score | -0.002311 (rank : 174) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WXS8, Q8TE55, Q8TEY8 | Gene names | ADAMTS14 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-14 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 14) (ADAM-TS 14) (ADAM-TS14). | |||||
|
CO5A3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 165) | NC score | -0.001094 (rank : 169) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
CO9A3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 166) | NC score | -0.001798 (rank : 172) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
EGR2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 167) | NC score | -0.009890 (rank : 179) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11161, Q8IV26, Q9UNA6 | Gene names | EGR2, KROX20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein) (AT591). | |||||
|
IRX2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 168) | NC score | -0.001930 (rank : 173) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZI1, Q7Z2I7 | Gene names | IRX2, IRXA2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
KRHB6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 169) | NC score | -0.004206 (rank : 177) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97861, Q3TTV9 | Gene names | Krthb6, Krt2-10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cuticular Hb6 (Hair keratin, type II Hb6). | |||||
|
MAML3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 170) | NC score | 0.000690 (rank : 164) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
NR4A3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 171) | NC score | -0.001514 (rank : 171) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZB6, Q9QZB5 | Gene names | Nr4a3, Tec | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A3 (Orphan nuclear receptor TEC) (Translocated in extraskeletal chondrosarcoma). | |||||
|
SN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 172) | NC score | -0.002873 (rank : 175) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62230, O55216, Q62228, Q62229 | Gene names | Sn, Sa | |||
|
Domain Architecture |

|
|||||
| Description | Sialoadhesin precursor (Sialic acid-binding Ig-like lectin-1) (Siglec- 1) (Sheep erythrocyte receptor) (SER) (CD169 antigen). | |||||
|
ZN446_HUMAN
|
||||||
| θ value | 8.99809 (rank : 173) | NC score | -0.008697 (rank : 178) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NWS9 | Gene names | ZNF446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 446. | |||||
|
ZYX_MOUSE
|
||||||
| θ value | 8.99809 (rank : 174) | NC score | 0.004189 (rank : 157) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62523, P70461 | Gene names | Zyx | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin. | |||||
|
CSTN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.397582 (rank : 143) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | Q99JH7 | Gene names | Clstn3, Cs3, Cstn3 | |||
|
Domain Architecture |
|
|||||
| Description | Calsyntenin-3 precursor. | |||||
|
EMR4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.052180 (rank : 144) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZE5, Q3U1I7, Q8BYX0, Q8VIM3 | Gene names | Emr4 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 4 precursor (EGF-like module-containing mucin-like receptor EMR4) (F4/80-like-receptor) (Seven-span membrane protein FIRE). | |||||
|
GP133_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.051962 (rank : 145) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6QNK2, Q6ZMQ1, Q7Z7M2, Q86SM4 | Gene names | GPR133, PGR25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 133 precursor (G-protein coupled receptor PGR25). | |||||
|
GP144_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.051884 (rank : 146) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z7M1, Q86SL4, Q8NH12 | Gene names | GPR144, PGR24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 144 (G-protein coupled receptor PGR24). | |||||
|
GPR98_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.051726 (rank : 147) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXG9, O75171, Q9H0X5, Q9UL61 | Gene names | GPR98, KIAA0686, MASS1, VLGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1 homolog) (Very large G-protein coupled receptor 1) (Usher syndrome type-2C protein). | |||||
|
GPR98_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.050481 (rank : 148) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VHN7, Q6ZQ69, Q810D2, Q810D3, Q91ZS0, Q91ZS1 | Gene names | Gpr98, Kiaa0686, Mass1, Vlgr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1) (Very large G-protein coupled receptor 1) (Neurepin). | |||||
|
DSG1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 174 | |
| SwissProt Accessions | Q02413 | Gene names | DSG1 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-1 precursor (Desmosomal glycoprotein 1) (DG1) (DGI) (Pemphigus foliaceus antigen). | |||||
|
DSG1B_MOUSE
|
||||||
| NC score | 0.998114 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q7TSF1, Q7TQ60 | Gene names | Dsg1b, Dsg5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-1 beta precursor (Dsg1-beta) (Desmoglein-5). | |||||
|
DSG1C_MOUSE
|
||||||
| NC score | 0.998095 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q7TSF0, Q7TQ61 | Gene names | Dsg1c, Dsg6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-1 gamma precursor (Dsg1-gamma) (Desmoglein-6). | |||||
|
DSG1A_MOUSE
|
||||||
| NC score | 0.998042 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q61495, Q8CE03 | Gene names | Dsg1a, Dsg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-1 alpha precursor (Dsg1-alpha) (Desmoglein-1) (Desmosomal glycoprotein I) (DG1) (DGI). | |||||
|
DSG4_MOUSE
|
||||||
| NC score | 0.987125 (rank : 5) | θ value | 1.2777e-171 (rank : 6) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q7TMD7 | Gene names | Dsg4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-4 precursor. | |||||
|
DSG3_MOUSE
|
||||||
| NC score | 0.986925 (rank : 6) | θ value | 1.24331e-150 (rank : 8) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | O35902, Q8CB02, Q8CE48 | Gene names | Dsg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-3 precursor (130 kDa pemphigus vulgaris antigen homolog). | |||||
|
DSG3_HUMAN
|
||||||
| NC score | 0.986793 (rank : 7) | θ value | 1.56915e-153 (rank : 7) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P32926 | Gene names | DSG3 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-3 precursor (130 kDa pemphigus vulgaris antigen) (PVA). | |||||
|
DSG4_HUMAN
|
||||||
| NC score | 0.986640 (rank : 8) | θ value | 5.18743e-173 (rank : 5) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q86SJ6, Q6Y9L9, Q8IXV4 | Gene names | DSG4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Desmoglein-4 precursor. | |||||
|
DSG2_HUMAN
|
||||||
| NC score | 0.978678 (rank : 9) | θ value | 4.29305e-135 (rank : 9) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q14126 | Gene names | DSG2 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-2 precursor (HDGC). | |||||
|
DSG2_MOUSE
|
||||||
| NC score | 0.977453 (rank : 10) | θ value | 7.63087e-108 (rank : 10) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | O55111, Q8K069, Q8R517 | Gene names | Dsg2 | |||
|
Domain Architecture |

|
|||||
| Description | Desmoglein-2 precursor. | |||||
|
DSC2_MOUSE
|
||||||
| NC score | 0.954452 (rank : 11) | θ value | 7.45998e-71 (rank : 11) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | P55292, Q64734 | Gene names | Dsc2, Dsc3 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-2 precursor (Epithelial type 2 desmocollin). | |||||
|
DSC3_HUMAN
|
||||||
| NC score | 0.952350 (rank : 12) | θ value | 3.83135e-67 (rank : 13) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q14574, Q14200, Q9HAZ9 | Gene names | DSC3, DSC4 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-3 precursor (Desmocollin-4) (HT-CP). | |||||
|
DSC2_HUMAN
|
||||||
| NC score | 0.951944 (rank : 13) | θ value | 4.83544e-70 (rank : 12) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q02487 | Gene names | DSC2, DSC3 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-2 precursor (Desmosomal glycoprotein II and III) (Desmocollin-3). | |||||
|
DSC3_MOUSE
|
||||||
| NC score | 0.951456 (rank : 14) | θ value | 1.45591e-66 (rank : 14) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55850, O55110, O55122 | Gene names | Dsc3 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-3 precursor. | |||||
|
CAD26_MOUSE
|
||||||
| NC score | 0.949962 (rank : 15) | θ value | 6.58091e-43 (rank : 30) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P59862 | Gene names | Cdh26 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-like protein 26 precursor. | |||||
|
CADH1_MOUSE
|
||||||
| NC score | 0.949902 (rank : 16) | θ value | 1.45929e-58 (rank : 15) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | P09803, Q61377 | Gene names | Cdh1 | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial-cadherin precursor (E-cadherin) (Uvomorulin) (Cadherin-1) (ARC-1) (CD324 antigen) [Contains: E-Cad/CTF1; E-Cad/CTF2; E- Cad/CTF3]. | |||||
|
CAD26_HUMAN
|
||||||
| NC score | 0.948567 (rank : 17) | θ value | 2.85459e-46 (rank : 29) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q8IXH8, Q8TCH3, Q9BQN4, Q9NRU1 | Gene names | CDH26 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-like protein 26 precursor (Cadherin-like protein VR20). | |||||
|
DSC1_MOUSE
|
||||||
| NC score | 0.948438 (rank : 18) | θ value | 6.13105e-57 (rank : 19) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55849 | Gene names | Dsc1 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-1 precursor. | |||||
|
CADH1_HUMAN
|
||||||
| NC score | 0.948394 (rank : 19) | θ value | 5.54529e-58 (rank : 18) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | P12830, Q13799, Q14216, Q15855, Q16194, Q4PJ14 | Gene names | CDH1, CDHE, UVO | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial-cadherin precursor (E-cadherin) (Uvomorulin) (Cadherin-1) (CAM 120/80) (CD324 antigen) [Contains: E-Cad/CTF1; E-Cad/CTF2; E- Cad/CTF3]. | |||||
|
DSC1_HUMAN
|
||||||
| NC score | 0.948065 (rank : 20) | θ value | 3.36421e-55 (rank : 21) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q08554 | Gene names | DSC1 | |||
|
Domain Architecture |

|
|||||
| Description | Desmocollin-1 precursor (Desmosomal glycoprotein 2/3) (DG2/DG3). | |||||
|
CAD13_HUMAN
|
||||||
| NC score | 0.947284 (rank : 21) | θ value | 3.60269e-49 (rank : 27) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55290, Q8TBX3 | Gene names | CDH13, CDHH | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-13 precursor (Truncated-cadherin) (T-cadherin) (T-cad) (Heart-cadherin) (H-cadherin) (P105). | |||||
|
CAD13_MOUSE
|
||||||
| NC score | 0.946789 (rank : 22) | θ value | 3.60269e-49 (rank : 28) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9WTR5 | Gene names | Cdh13 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-13 precursor (Truncated-cadherin) (T-cadherin) (T-cad) (Heart-cadherin) (H-cadherin). | |||||
|
CADH3_MOUSE
|
||||||
| NC score | 0.945965 (rank : 23) | θ value | 3.1488e-53 (rank : 23) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P10287, Q61465 | Gene names | Cdh3, Cdhp | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-3 precursor (Placental-cadherin) (P-cadherin). | |||||
|
CAD15_MOUSE
|
||||||
| NC score | 0.945523 (rank : 24) | θ value | 7.25919e-50 (rank : 26) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | P33146, Q9QYZ7 | Gene names | Cdh15, Cdh14 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-15 precursor (Muscle-cadherin) (M-cadherin) (Cadherin-14). | |||||
|
CADH3_HUMAN
|
||||||
| NC score | 0.945494 (rank : 25) | θ value | 9.16162e-53 (rank : 24) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P22223 | Gene names | CDH3, CDHP | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-3 precursor (Placental-cadherin) (P-cadherin). | |||||
|
CADH2_HUMAN
|
||||||
| NC score | 0.945199 (rank : 26) | θ value | 2.32979e-56 (rank : 20) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P19022, Q14923 | Gene names | CDH2, CDHN, NCAD | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-2 precursor (Neural-cadherin) (N-cadherin) (CDw325 antigen). | |||||
|
CAD15_HUMAN
|
||||||
| NC score | 0.944661 (rank : 27) | θ value | 1.11993e-50 (rank : 25) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | P55291 | Gene names | CDH15, CDH14, CDH3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-15 precursor (Muscle-cadherin) (M-cadherin) (Cadherin-14). | |||||
|
CADH4_MOUSE
|
||||||
| NC score | 0.944519 (rank : 28) | θ value | 3.25095e-58 (rank : 16) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P39038 | Gene names | Cdh4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
CADH4_HUMAN
|
||||||
| NC score | 0.944225 (rank : 29) | θ value | 4.24587e-58 (rank : 17) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55283, Q2M208, Q5VZ44, Q9BZ05 | Gene names | CDH4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
CADH2_MOUSE
|
||||||
| NC score | 0.944034 (rank : 30) | θ value | 3.71957e-54 (rank : 22) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P15116, Q64260 | Gene names | Cdh2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-2 precursor (Neural-cadherin) (N-cadherin) (CDw325 antigen). | |||||
|
CAD17_HUMAN
|
||||||
| NC score | 0.923869 (rank : 31) | θ value | 8.06329e-33 (rank : 39) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q12864, Q15336 | Gene names | CDH17 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-17 precursor (Liver-intestine-cadherin) (LI-cadherin) (Intestinal peptide-associated transporter HPT-1). | |||||
|
CAD17_MOUSE
|
||||||
| NC score | 0.922484 (rank : 32) | θ value | 8.06329e-33 (rank : 40) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9R100 | Gene names | Cdh17 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-17 precursor (Liver-intestine-cadherin) (LI-cadherin) (BILL- cadherin) (P130). | |||||
|
CADH5_HUMAN
|
||||||
| NC score | 0.908323 (rank : 33) | θ value | 1.98606e-31 (rank : 43) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P33151 | Gene names | CDH5 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-5 precursor (Vascular endothelial-cadherin) (VE-cadherin) (7B4 antigen) (CD144 antigen). | |||||
|
CADH5_MOUSE
|
||||||
| NC score | 0.907710 (rank : 34) | θ value | 9.8567e-31 (rank : 44) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55284, O35542 | Gene names | Cdh5 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-5 precursor (Vascular endothelial-cadherin) (VE-cadherin) (CD144 antigen). | |||||
|
CAD12_HUMAN
|
||||||
| NC score | 0.907613 (rank : 35) | θ value | 5.21438e-40 (rank : 31) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55289 | Gene names | CDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-12 precursor (Brain-cadherin) (BR-cadherin) (N-cadherin 2) (Neural type cadherin 2). | |||||
|
CAD18_HUMAN
|
||||||
| NC score | 0.907323 (rank : 36) | θ value | 1.73584e-35 (rank : 35) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q13634 | Gene names | CDH18, CDH14 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-18 precursor (Cadherin-14). | |||||
|
CADH7_HUMAN
|
||||||
| NC score | 0.905219 (rank : 37) | θ value | 3.06405e-32 (rank : 42) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9ULB5, Q9H157 | Gene names | CDH7, CDH7L1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-7 precursor. | |||||
|
CADH6_HUMAN
|
||||||
| NC score | 0.904821 (rank : 38) | θ value | 3.16345e-37 (rank : 32) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55285, Q9BWS0 | Gene names | CDH6 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-6 precursor (Kidney-cadherin) (K-cadherin). | |||||
|
CAD11_HUMAN
|
||||||
| NC score | 0.904319 (rank : 39) | θ value | 5.96599e-36 (rank : 33) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55287, Q15065, Q15066, Q9UQ93, Q9UQ94 | Gene names | CDH11 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-11 precursor (Osteoblast-cadherin) (OB-cadherin) (OSF-4). | |||||
|
CAD11_MOUSE
|
||||||
| NC score | 0.904166 (rank : 40) | θ value | 7.79185e-36 (rank : 34) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55288 | Gene names | Cdh11, Cad-11 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-11 precursor (Osteoblast-cadherin) (OB-cadherin) (OSF-4). | |||||
|
CADH6_MOUSE
|
||||||
| NC score | 0.902960 (rank : 41) | θ value | 3.86705e-35 (rank : 36) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P97326, P70393 | Gene names | Cdh6 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-6 precursor (Kidney-cadherin) (K-cadherin). | |||||
|
CAD20_HUMAN
|
||||||
| NC score | 0.902750 (rank : 42) | θ value | 4.27553e-34 (rank : 38) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9HBT6 | Gene names | CDH20, CDH7L3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-20 precursor. | |||||
|
CAD10_HUMAN
|
||||||
| NC score | 0.902624 (rank : 43) | θ value | 2.50655e-34 (rank : 37) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y6N8, Q9ULB3 | Gene names | CDH10 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-10 precursor (T2-cadherin). | |||||
|
CAD24_HUMAN
|
||||||
| NC score | 0.902512 (rank : 44) | θ value | 3.06405e-32 (rank : 41) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q86UP0, Q86UP1, Q9NT84 | Gene names | CDH24, CDH11L | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-24 precursor. | |||||
|
CAD22_MOUSE
|
||||||
| NC score | 0.900711 (rank : 45) | θ value | 2.42779e-29 (rank : 46) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9WTP5 | Gene names | Cdh22 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-22 precursor (Pituitary and brain cadherin) (PB-cadherin). | |||||
|
CAD22_HUMAN
|
||||||
| NC score | 0.898359 (rank : 46) | θ value | 5.06226e-27 (rank : 50) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9UJ99, O43205 | Gene names | CDH22, C20orf25 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-22 precursor (Pituitary and brain cadherin) (PB-cadherin). | |||||
|
CADH9_HUMAN
|
||||||
| NC score | 0.898220 (rank : 47) | θ value | 3.74554e-30 (rank : 45) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9ULB4 | Gene names | CDH9 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-9 precursor. | |||||
|
CAD19_HUMAN
|
||||||
| NC score | 0.897919 (rank : 48) | θ value | 2.05525e-28 (rank : 48) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9H159 | Gene names | CDH19, CDH7L2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-19 precursor. | |||||
|
CADH8_HUMAN
|
||||||
| NC score | 0.897029 (rank : 49) | θ value | 1.57365e-28 (rank : 47) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P55286, Q9ULB2 | Gene names | CDH8 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-8 precursor. | |||||
|
CADH8_MOUSE
|
||||||
| NC score | 0.896635 (rank : 50) | θ value | 3.50572e-28 (rank : 49) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | P97291 | Gene names | Cdh8 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-8 precursor. | |||||
|
CAD16_MOUSE
|
||||||
| NC score | 0.893444 (rank : 51) | θ value | 2.13179e-17 (rank : 94) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | O88338, Q9JLZ5 | Gene names | Cdh16 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-16 precursor (Kidney-specific cadherin) (Ksp-cadherin). | |||||
|
CAD16_HUMAN
|
||||||
| NC score | 0.891196 (rank : 52) | θ value | 3.88503e-19 (rank : 72) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | O75309, Q6UW93 | Gene names | CDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-16 precursor (Kidney-specific cadherin) (Ksp-cadherin). | |||||
|
PCDLK_HUMAN
|
||||||
| NC score | 0.848945 (rank : 53) | θ value | 4.02038e-16 (rank : 110) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9BYE9, Q9NXP8 | Gene names | PCLKC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin LKC precursor (PC-LKC). | |||||
|
CAD23_HUMAN
|
||||||
| NC score | 0.838205 (rank : 54) | θ value | 9.87957e-23 (rank : 54) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9H251, Q6UWW1, Q9H4K9 | Gene names | CDH23, KIAA1774 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
CAD23_MOUSE
|
||||||
| NC score | 0.838060 (rank : 55) | θ value | 9.87957e-23 (rank : 55) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q99PF4, Q99NH1, Q9D4N9 | Gene names | Cdh23 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-23 precursor (Otocadherin). | |||||
|
PCD15_MOUSE
|
||||||
| NC score | 0.825564 (rank : 56) | θ value | 2.43908e-13 (rank : 128) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q99PJ1 | Gene names | Pcdh15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
PCD15_HUMAN
|
||||||
| NC score | 0.823137 (rank : 57) | θ value | 1.86753e-13 (rank : 127) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
PCD16_HUMAN
|
||||||
| NC score | 0.821439 (rank : 58) | θ value | 1.24977e-17 (rank : 89) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q96JQ0, O15098 | Gene names | DCHS1, CDH19, FIB1, KIAA1773, PCDH16 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-16 precursor (Dachsous 1) (Cadherin-19) (Fibroblast cadherin 1). | |||||
|
PCDH1_HUMAN
|
||||||
| NC score | 0.814427 (rank : 59) | θ value | 7.82807e-20 (rank : 65) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q08174 | Gene names | PCDH1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-1 precursor (Protocadherin-42) (PC42) (Cadherin-like protein 1). | |||||
|
PCDH7_HUMAN
|
||||||
| NC score | 0.808551 (rank : 60) | θ value | 1.24977e-17 (rank : 91) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | O60245, O60246, O60247 | Gene names | PCDH7, BHPCDH | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-7 precursor (Brain-heart protocadherin) (BH-Pcdh). | |||||
|
PCD20_HUMAN
|
||||||
| NC score | 0.807299 (rank : 61) | θ value | 8.10077e-17 (rank : 102) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q8N6Y1, Q8NDN4, Q9NRT9 | Gene names | PCDH20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-20 precursor (Protocadherin-13). | |||||
|
PCD20_MOUSE
|
||||||
| NC score | 0.804928 (rank : 62) | θ value | 3.18553e-13 (rank : 130) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q8BIZ0, Q8BIV2 | Gene names | Pcdh20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-20 precursor. | |||||
|
PCDH9_HUMAN
|
||||||
| NC score | 0.804897 (rank : 63) | θ value | 1.38178e-16 (rank : 107) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9HC56 | Gene names | PCDH9 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-9 precursor. | |||||
|
PC11Y_HUMAN
|
||||||
| NC score | 0.803482 (rank : 64) | θ value | 2.27762e-19 (rank : 71) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q9BZA8, Q70LR6, Q70LR8, Q70LS0, Q70LS1, Q70LS2, Q70LS3, Q70LS4, Q70LS5, Q8WY34, Q9BZA9, Q9H4E1 | Gene names | PCDH11Y, PCDH11, PCDH22, PCDHY | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 Y-linked precursor (Protocadherin-11) (Protocadherin- 22) (Protocadherin on the Y chromosome) (PCDH-Y) (Protocadherin prostate cancer) (Protocadherin-PC). | |||||
|
PC11X_HUMAN
|
||||||
| NC score | 0.801854 (rank : 65) | θ value | 1.13037e-18 (rank : 77) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q9BZA7, Q2TJH0, Q2TJH1, Q2TJH3, Q5JVZ0, Q70LR8, Q70LS7, Q70LS8, Q70LS9, Q70LT7, Q70LT8, Q70LT9, Q70LU0, Q70LU1, Q96RV4, Q96RW0, Q9BZA6, Q9H4E0, Q9P2M0, Q9P2X5 | Gene names | PCDH11X, PCDH11, PCDHX | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-11 X-linked precursor (Protocadherin-11) (Protocadherin on the X chromosome) (PCDH-X) (Protocadherin-S). | |||||
|
FAT2_HUMAN
|
||||||
| NC score | 0.792674 (rank : 66) | θ value | 2.06002e-20 (rank : 60) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 507 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9NYQ8, O75091, Q9NSR7 | Gene names | FAT2, CDHF8, KIAA0811, MEGF1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin Fat 2 precursor (hFat2) (Multiple epidermal growth factor-like domains 1). | |||||
|
PCDC2_HUMAN
|
||||||
| NC score | 0.785667 (rank : 67) | θ value | 8.65492e-19 (rank : 76) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9Y5I4, Q9Y5F4 | Gene names | PCDHAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C2 precursor (PCDH-alpha-C2). | |||||
|
PCDGG_HUMAN
|
||||||
| NC score | 0.781978 (rank : 68) | θ value | 4.29542e-18 (rank : 86) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9UN71, O15099, Q9UN64 | Gene names | PCDHGB4, CDH20, FIB2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B4 precursor (PCDH-gamma-B4) (Cadherin-20) (Fibroblast cadherin 2). | |||||
|
PCD18_HUMAN
|
||||||
| NC score | 0.781309 (rank : 69) | θ value | 9.90251e-15 (rank : 123) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9HCL0 | Gene names | PCDH18, KIAA1562 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-18 precursor. | |||||
|
PCD12_MOUSE
|
||||||
| NC score | 0.780238 (rank : 70) | θ value | 1.16975e-15 (rank : 115) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | O55134 | Gene names | Pcdh12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
PCDGH_HUMAN
|
||||||
| NC score | 0.779776 (rank : 71) | θ value | 1.02238e-19 (rank : 67) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G0, Q9Y5C6 | Gene names | PCDHGB5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B5 precursor (PCDH-gamma-B5). | |||||
|
PCD12_HUMAN
|
||||||
| NC score | 0.779277 (rank : 72) | θ value | 2.13179e-17 (rank : 95) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q9NPG4, Q6UXB6, Q96KB8, Q9H7Y6, Q9H8E0 | Gene names | PCDH12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-12 precursor (Vascular cadherin-2) (Vascular endothelial cadherin-2) (VE-cadherin-2) (VE-cad-2). | |||||
|
PCD17_HUMAN
|
||||||
| NC score | 0.778697 (rank : 73) | θ value | 5.25075e-16 (rank : 111) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 140 | |
| SwissProt Accessions | O14917 | Gene names | PCDH17, PCDH68, PCH68 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-17 precursor (Protocadherin-68). | |||||
|
PCDBC_HUMAN
|
||||||
| NC score | 0.778417 (rank : 74) | θ value | 7.82807e-20 (rank : 64) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5F1 | Gene names | PCDHB12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 12 precursor (PCDH-beta12). | |||||
|
PCD10_HUMAN
|
||||||
| NC score | 0.778409 (rank : 75) | θ value | 1.058e-16 (rank : 103) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9P2E7 | Gene names | PCDH10, KIAA1400 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-10 precursor. | |||||
|
PCD19_MOUSE
|
||||||
| NC score | 0.778086 (rank : 76) | θ value | 2.69671e-12 (rank : 132) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q80TF3, Q8BL13 | Gene names | Pcdh19, Kiaa1313 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-19 precursor. | |||||
|
PCD19_HUMAN
|
||||||
| NC score | 0.778085 (rank : 77) | θ value | 4.16044e-13 (rank : 131) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q8TAB3, Q5JTG1, Q5JTG2, Q68DT7, Q9P2N3 | Gene names | PCDH19, KIAA1313 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-19 precursor. | |||||
|
PCDB6_HUMAN
|
||||||
| NC score | 0.777664 (rank : 78) | θ value | 1.02238e-19 (rank : 66) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5E3 | Gene names | PCDHB6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 6 precursor (PCDH-beta6). | |||||
|
PCDBG_HUMAN
|
||||||
| NC score | 0.777078 (rank : 79) | θ value | 1.33526e-19 (rank : 69) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9NRJ7, Q8IYD5, Q96SE9, Q9HCF1 | Gene names | PCDHB16, KIAA1621, PCDH3X | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 16 precursor (PCDH-beta16) (Protocadherin 3X). | |||||
|
PCDGD_HUMAN
|
||||||
| NC score | 0.776740 (rank : 80) | θ value | 4.29542e-18 (rank : 85) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G3, Q9Y5C8 | Gene names | PCDHGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B1 precursor (PCDH-gamma-B1). | |||||
|
PCDB7_HUMAN
|
||||||
| NC score | 0.775106 (rank : 81) | θ value | 1.63225e-17 (rank : 92) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5E2 | Gene names | PCDHB7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 7 precursor (PCDH-beta7). | |||||
|
PCDB9_HUMAN
|
||||||
| NC score | 0.774476 (rank : 82) | θ value | 3.28887e-18 (rank : 82) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5E1, Q9NRJ8 | Gene names | PCDHB9, PCDH3H | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 9 precursor (PCDH-beta9) (Protocadherin 3H). | |||||
|
PCDGK_HUMAN
|
||||||
| NC score | 0.774310 (rank : 83) | θ value | 6.62687e-19 (rank : 75) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9UN70, O60622, Q08192, Q9Y5C4 | Gene names | PCDHGC3, PCDH2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C3 precursor (PCDH-gamma-C3) (Protocadherin 43) (PC-43) (Protocadherin 2). | |||||
|
PCDBF_HUMAN
|
||||||
| NC score | 0.774133 (rank : 84) | θ value | 1.92812e-18 (rank : 81) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q9Y5E8, Q8IUX5 | Gene names | PCDHB15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 15 precursor (PCDH-beta15). | |||||
|
PCDB4_HUMAN
|
||||||
| NC score | 0.773874 (rank : 85) | θ value | 1.99529e-15 (rank : 118) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5E5 | Gene names | PCDHB4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 4 precursor (PCDH-beta4). | |||||
|
PCDBB_HUMAN
|
||||||
| NC score | 0.773545 (rank : 86) | θ value | 3.88503e-19 (rank : 73) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5F2 | Gene names | PCDHB11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 11 precursor (PCDH-beta11). | |||||
|
PCDB8_HUMAN
|
||||||
| NC score | 0.772936 (rank : 87) | θ value | 1.63225e-17 (rank : 93) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9UN66 | Gene names | PCDHB8, PCDH3I | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 8 precursor (PCDH-beta8) (Protocadherin 3I). | |||||
|
PCDGJ_HUMAN
|
||||||
| NC score | 0.772175 (rank : 88) | θ value | 6.41864e-14 (rank : 126) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5F8, Q9UN63 | Gene names | PCDHGB7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B7 precursor (PCDH-gamma-B7). | |||||
|
PCDG9_HUMAN
|
||||||
| NC score | 0.772005 (rank : 89) | θ value | 2.7842e-17 (rank : 98) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G4, Q9Y5C9 | Gene names | PCDHGA9 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A9 precursor (PCDH-gamma-A9). | |||||
|
PCDAB_HUMAN
|
||||||
| NC score | 0.771964 (rank : 90) | θ value | 1.09232e-21 (rank : 56) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 211 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9Y5I1, O75279 | Gene names | PCDHA11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 11 precursor (PCDH-alpha11). | |||||
|
PCDBE_HUMAN
|
||||||
| NC score | 0.771810 (rank : 91) | θ value | 9.56915e-18 (rank : 88) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q9Y5E9 | Gene names | PCDHB14 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 14 precursor (PCDH-beta14). | |||||
|
PCDGF_HUMAN
|
||||||
| NC score | 0.771569 (rank : 92) | θ value | 1.13037e-18 (rank : 80) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G1, Q9Y5C7 | Gene names | PCDHGB3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B3 precursor (PCDH-gamma-B3). | |||||
|
PCDA2_HUMAN
|
||||||
| NC score | 0.771290 (rank : 93) | θ value | 1.13037e-18 (rank : 78) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9Y5H9, O75287, Q9BTV3 | Gene names | PCDHA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 2 precursor (PCDH-alpha2). | |||||
|
PCDGB_HUMAN
|
||||||
| NC score | 0.771080 (rank : 94) | θ value | 4.29542e-18 (rank : 84) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5H2, Q9Y5D8, Q9Y5D9 | Gene names | PCDHGA11 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A11 precursor (PCDH-gamma-A11). | |||||
|
PCDG2_HUMAN
|
||||||
| NC score | 0.770776 (rank : 95) | θ value | 1.5773e-20 (rank : 58) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 151 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5H1, Q9Y5D5 | Gene names | PCDHGA2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A2 precursor (PCDH-gamma-A2). | |||||
|
PCDBD_HUMAN
|
||||||
| NC score | 0.770610 (rank : 96) | θ value | 1.99529e-15 (rank : 119) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5F0 | Gene names | PCDHB13 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 13 precursor (PCDH-beta13). | |||||
|
PCDB5_HUMAN
|
||||||
| NC score | 0.770607 (rank : 97) | θ value | 1.33526e-19 (rank : 68) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q9Y5E4, Q9UFU9 | Gene names | PCDHB5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 5 precursor (PCDH-beta5). | |||||
|
PCDGI_HUMAN
|
||||||
| NC score | 0.769422 (rank : 98) | θ value | 3.40345e-15 (rank : 122) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5F9, Q9Y5C5 | Gene names | PCDHGB6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B6 precursor (PCDH-gamma-B6). | |||||
|
PCDGL_HUMAN
|
||||||
| NC score | 0.769119 (rank : 99) | θ value | 2.43908e-13 (rank : 129) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 157 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5F7, Q9Y5C3 | Gene names | PCDHGC4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C4 precursor (PCDH-gamma-C4). | |||||
|
PCDG8_HUMAN
|
||||||
| NC score | 0.769087 (rank : 100) | θ value | 4.74913e-17 (rank : 101) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G5, O15039 | Gene names | PCDHGA8, KIAA0327 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A8 precursor (PCDH-gamma-A8). | |||||
|
PCDA1_HUMAN
|
||||||
| NC score | 0.768596 (rank : 101) | θ value | 2.69047e-20 (rank : 61) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9Y5I3, O75288, Q9NRT7 | Gene names | PCDHA1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 1 precursor (PCDH-alpha1). | |||||
|
PCDG6_HUMAN
|
||||||
| NC score | 0.767988 (rank : 102) | θ value | 3.88503e-19 (rank : 74) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G7, Q9Y5D1 | Gene names | PCDHGA6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A6 precursor (PCDH-gamma-A6). | |||||
|
PCDB2_HUMAN
|
||||||
| NC score | 0.767929 (rank : 103) | θ value | 4.74913e-17 (rank : 100) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q9Y5E7 | Gene names | PCDHB2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 2 precursor (PCDH-beta2). | |||||
|
PCDGC_HUMAN
|
||||||
| NC score | 0.767694 (rank : 104) | θ value | 1.13037e-18 (rank : 79) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | O60330, O15100, Q6UW70, Q9Y5D7 | Gene names | PCDHGA12, CDH21, FIB3, KIAA0588 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A12 precursor (PCDH-gamma-A12) (Cadherin-21) (Fibroblast cadherin 3). | |||||
|
PCDG1_HUMAN
|
||||||
| NC score | 0.767657 (rank : 105) | θ value | 7.32683e-18 (rank : 87) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 158 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5H4, Q9Y5D6 | Gene names | PCDHGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A1 precursor (PCDH-gamma-A1). | |||||
|
PCDG3_HUMAN
|
||||||
| NC score | 0.767648 (rank : 106) | θ value | 4.02038e-16 (rank : 109) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 155 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5H0, Q9Y5D4 | Gene names | PCDHGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A3 precursor (PCDH-gamma-A3). | |||||
|
PCDBA_HUMAN
|
||||||
| NC score | 0.767311 (rank : 107) | θ value | 1.058e-16 (rank : 104) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9UN67, Q96T99 | Gene names | PCDHB10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 10 precursor (PCDH-beta10). | |||||
|
PCDA5_HUMAN
|
||||||
| NC score | 0.767219 (rank : 108) | θ value | 2.69047e-20 (rank : 62) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9Y5H7, O75284, Q8N4R3 | Gene names | PCDHA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 5 precursor (PCDH-alpha5). | |||||
|
PCDGE_HUMAN
|
||||||
| NC score | 0.767074 (rank : 109) | θ value | 1.52774e-15 (rank : 116) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G2, Q9UN65 | Gene names | PCDHGB2 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma B2 precursor (PCDH-gamma-B2). | |||||
|
PCDB3_HUMAN
|
||||||
| NC score | 0.766847 (rank : 110) | θ value | 5.25075e-16 (rank : 113) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5E6 | Gene names | PCDHB3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 3 precursor (PCDH-beta3). | |||||
|
PCDA4_MOUSE
|
||||||
| NC score | 0.766641 (rank : 111) | θ value | 1.24977e-17 (rank : 90) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 146 | |
| SwissProt Accessions | O88689, Q3UEX3, Q6PAM9, Q8K487, Q8K489 | Gene names | Pcdha4, Cnr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin alpha 4 precursor (PCDH-alpha4). | |||||
|
PCDAD_HUMAN
|
||||||
| NC score | 0.766502 (rank : 112) | θ value | 2.7842e-17 (rank : 97) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9Y5I0, O75277 | Gene names | PCDHA13 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 13 precursor (PCDH-alpha13). | |||||
|
PCDG5_HUMAN
|
||||||
| NC score | 0.766132 (rank : 113) | θ value | 1.80466e-16 (rank : 108) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G8, Q9Y5D2 | Gene names | PCDHGA5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A5 precursor (PCDH-gamma-A5). | |||||
|
PCDB1_HUMAN
|
||||||
| NC score | 0.766052 (rank : 114) | θ value | 8.95645e-16 (rank : 114) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5F3 | Gene names | PCDHB1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin beta 1 precursor (PCDH-beta1). | |||||
|
PCDG7_HUMAN
|
||||||
| NC score | 0.765599 (rank : 115) | θ value | 3.63628e-17 (rank : 99) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G6, Q9Y5D0 | Gene names | PCDHGA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A7 precursor (PCDH-gamma-A7). | |||||
|
PCDGA_HUMAN
|
||||||
| NC score | 0.764292 (rank : 116) | θ value | 3.28887e-18 (rank : 83) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5H3, Q9Y5E0 | Gene names | PCDHGA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A10 precursor (PCDH-gamma-A10). | |||||
|
PCDGM_HUMAN
|
||||||
| NC score | 0.763012 (rank : 117) | θ value | 1.52774e-15 (rank : 117) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 141 | |
| SwissProt Accessions | Q9Y5F6, Q9Y5C2 | Gene names | PCDHGC5 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma C5 precursor (PCDH-gamma-C5). | |||||
|
PCDA8_HUMAN
|
||||||
| NC score | 0.761965 (rank : 118) | θ value | 1.38178e-16 (rank : 105) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9Y5H6, O75281 | Gene names | PCDHA8 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 8 precursor (PCDH-alpha8). | |||||
|
PCDG4_HUMAN
|
||||||
| NC score | 0.761583 (rank : 119) | θ value | 1.99529e-15 (rank : 120) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | Q9Y5G9, Q9Y5D3 | Gene names | PCDHGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin gamma A4 precursor (PCDH-gamma-A4). | |||||
|
PCDA6_HUMAN
|
||||||
| NC score | 0.760830 (rank : 120) | θ value | 5.25075e-16 (rank : 112) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9UN73, O75283, Q9NRT8 | Gene names | PCDHA6 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 6 precursor (PCDH-alpha6). | |||||
|
PCDA3_HUMAN
|
||||||
| NC score | 0.760675 (rank : 121) | θ value | 2.13179e-17 (rank : 96) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9Y5H8, O75286 | Gene names | PCDHA3 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 3 precursor (PCDH-alpha3). | |||||
|
PCDC1_HUMAN
|
||||||
| NC score | 0.758573 (rank : 122) | θ value | 1.74796e-11 (rank : 133) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9H158, Q9Y5F5, Q9Y5I5 | Gene names | PCDHAC1 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha C1 precursor (PCDH-alpha-C1). | |||||
|
PCDAC_HUMAN
|
||||||
| NC score | 0.758059 (rank : 123) | θ value | 2.88119e-14 (rank : 125) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9UN75, O75278 | Gene names | PCDHA12 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 12 precursor (PCDH-alpha12). | |||||
|
PCDA9_HUMAN
|
||||||
| NC score | 0.757518 (rank : 124) | θ value | 3.40345e-15 (rank : 121) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9Y5H5, O15053 | Gene names | PCDHA9, KIAA0345 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 9 precursor (PCDH-alpha9). | |||||
|
PCDA4_HUMAN
|
||||||
| NC score | 0.755181 (rank : 125) | θ value | 2.28291e-11 (rank : 134) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9UN74, O75285 | Gene names | PCDHA4 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 4 precursor (PCDH-alpha4). | |||||
|
PCDA7_HUMAN
|
||||||
| NC score | 0.753949 (rank : 126) | θ value | 2.88119e-14 (rank : 124) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9UN72, O75282 | Gene names | PCDHA7 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 7 precursor (PCDH-alpha7). | |||||
|
PCDAA_HUMAN
|
||||||
| NC score | 0.753320 (rank : 127) | θ value | 1.38178e-16 (rank : 106) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9Y5I2, O75280, Q9NRU2 | Gene names | PCDHA10 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin alpha 10 precursor (PCDH-alpha10). | |||||
|
FATH_HUMAN
|
||||||
| NC score | 0.745642 (rank : 128) | θ value | 4.73814e-25 (rank : 52) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 570 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q14517 | Gene names | FAT | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-related tumor suppressor homolog precursor (Protein fat homolog). | |||||
|
PCDH8_HUMAN
|
||||||
| NC score | 0.740786 (rank : 129) | θ value | 8.97725e-08 (rank : 135) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 142 | |
| SwissProt Accessions | O95206, Q8IYE9, Q96SF1 | Gene names | PCDH8 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-8 precursor (Arcadlin). | |||||
|
CELR2_MOUSE
|
||||||
| NC score | 0.700921 (rank : 130) | θ value | 1.74391e-19 (rank : 70) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
CELR3_MOUSE
|
||||||
| NC score | 0.699108 (rank : 131) | θ value | 2.06002e-20 (rank : 59) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 687 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q91ZI0, Q9ESD0 | Gene names | Celsr3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor. | |||||
|
CELR3_HUMAN
|
||||||
| NC score | 0.697970 (rank : 132) | θ value | 1.2077e-20 (rank : 57) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 680 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9NYQ7, O75092 | Gene names | CELSR3, CDHF11, EGFL1, FMI1, KIAA0812, MEGF2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 3 precursor (Flamingo homolog 1) (hFmi1) (Multiple epidermal growth factor-like domains 2) (Epidermal growth factor-like 1). | |||||
|
CELR1_MOUSE
|
||||||
| NC score | 0.696722 (rank : 133) | θ value | 7.56453e-23 (rank : 53) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | O35161 | Gene names | Celsr1 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor. | |||||
|
CELR1_HUMAN
|
||||||
| NC score | 0.696100 (rank : 134) | θ value | 2.12685e-25 (rank : 51) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 613 | Shared Neighborhood Hits | 143 | |
| SwissProt Accessions | Q9NYQ6, O95722, Q5TH47, Q9BWQ5, Q9Y506, Q9Y526 | Gene names | CELSR1, CDHF9, FMI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 1 precursor (Flamingo homolog 2) (hFmi2). | |||||
|
CELR2_HUMAN
|
||||||
| NC score | 0.694571 (rank : 135) | θ value | 3.51386e-20 (rank : 63) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 144 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.600883 (rank : 136) | θ value | 0.0193708 (rank : 141) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 141 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
MUCDL_HUMAN
|
||||||
| NC score | 0.548875 (rank : 137) | θ value | 0.125558 (rank : 142) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 145 | |
| SwissProt Accessions | Q9HBB8, Q9H746, Q9HAU3, Q9HBB5, Q9HBB6, Q9HBB7, Q9NX86, Q9NXI9 | Gene names | MUCDHL | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
CSTN1_MOUSE
|
||||||
| NC score | 0.515880 (rank : 138) | θ value | 1.87187e-05 (rank : 136) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q9EPL2 | Gene names | Clstn1, Cs1, Cstn1 | |||
|
Domain Architecture |

|
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
CSTN2_MOUSE
|
||||||
| NC score | 0.495183 (rank : 139) | θ value | 0.00035302 (rank : 139) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | Q9ER65 | Gene names | Clstn2, Cs2, Cstn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calsyntenin-2 precursor. | |||||
|
CSTN1_HUMAN
|
||||||
| NC score | 0.491045 (rank : 140) | θ value | 0.000158464 (rank : 137) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 138 | |
| SwissProt Accessions | O94985 | Gene names | CLSTN1, CS1, KIAA0911 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calsyntenin-1 precursor. | |||||
|
CSTN2_HUMAN
|
||||||
| NC score | 0.491022 (rank : 141) | θ value | 0.000270298 (rank : 138) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 136 | |
| SwissProt Accessions | Q9H4D0, Q9BSS0 | Gene names | CLSTN2, CS2 | |||
|
Domain Architecture |

|
|||||
| Description | Calsyntenin-2 precursor. | |||||
|
CSTN3_HUMAN
|
||||||
| NC score | 0.426566 (rank : 142) | θ value | 0.00665767 (rank : 140) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 137 | |
| SwissProt Accessions | Q9BQT9, O94831 | Gene names | CLSTN3, CS3, KIAA0726 | |||
|
Domain Architecture |

|
|||||
| Description | Calsyntenin-3 precursor. | |||||
|
CSTN3_MOUSE
|
||||||
| NC score | 0.397582 (rank : 143) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 132 | |
| SwissProt Accessions | Q99JH7 | Gene names | Clstn3, Cs3, Cstn3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsyntenin-3 precursor. | |||||
|
EMR4_MOUSE
|
||||||
| NC score | 0.052180 (rank : 144) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91ZE5, Q3U1I7, Q8BYX0, Q8VIM3 | Gene names | Emr4 | |||
|
Domain Architecture |

|
|||||
| Description | EGF-like module-containing mucin-like hormone receptor-like 4 precursor (EGF-like module-containing mucin-like receptor EMR4) (F4/80-like-receptor) (Seven-span membrane protein FIRE). | |||||
|
GP133_HUMAN
|
||||||
| NC score | 0.051962 (rank : 145) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6QNK2, Q6ZMQ1, Q7Z7M2, Q86SM4 | Gene names | GPR133, PGR25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 133 precursor (G-protein coupled receptor PGR25). | |||||
|
GP144_HUMAN
|
||||||
| NC score | 0.051884 (rank : 146) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q7Z7M1, Q86SL4, Q8NH12 | Gene names | GPR144, PGR24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 144 (G-protein coupled receptor PGR24). | |||||
|
GPR98_HUMAN
|
||||||
| NC score | 0.051726 (rank : 147) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8WXG9, O75171, Q9H0X5, Q9UL61 | Gene names | GPR98, KIAA0686, MASS1, VLGR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1 homolog) (Very large G-protein coupled receptor 1) (Usher syndrome type-2C protein). | |||||
|
GPR98_MOUSE
|
||||||
| NC score | 0.050481 (rank : 148) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q8VHN7, Q6ZQ69, Q810D2, Q810D3, Q91ZS0, Q91ZS1 | Gene names | Gpr98, Kiaa0686, Mass1, Vlgr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G-protein coupled receptor 98 precursor (Monogenic audiogenic seizure susceptibility protein 1) (Very large G-protein coupled receptor 1) (Neurepin). | |||||
|
CSPG2_MOUSE
|
||||||
| NC score | 0.031688 (rank : 149) | θ value | 4.03905 (rank : 153) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 711 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62059, Q62058, Q9CUU0 | Gene names | Cspg2 | |||
|
Domain Architecture |

|
|||||
| Description | Versican core protein precursor (Large fibroblast proteoglycan) (Chondroitin sulfate proteoglycan core protein 2) (PG-M). | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.016714 (rank : 150) | θ value | 4.03905 (rank : 151) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CBP_HUMAN
|
||||||
| NC score | 0.009357 (rank : 151) | θ value | 1.38821 (rank : 147) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 941 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q92793, O00147, Q16376 | Gene names | CREBBP, CBP | |||
|
Domain Architecture |

|
|||||
| Description | CREB-binding protein (EC 2.3.1.48). | |||||
|
PER1_HUMAN
|
||||||
| NC score | 0.008400 (rank : 152) | θ value | 0.279714 (rank : 144) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O15534 | Gene names | PER1, KIAA0482, PER, RIGUI | |||
|
Domain Architecture |

|
|||||
| Description | Period circadian protein 1 (Circadian pacemaker protein Rigui) (hPER). | |||||
|
PXDC1_HUMAN
|
||||||
| NC score | 0.007429 (rank : 153) | θ value | 1.81305 (rank : 149) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IUK5, Q5QCZ7, Q5QCZ8, Q5QCZ9, Q9HCT9 | Gene names | PLXDC1, TEM3, TEM7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Plexin domain-containing protein 1 precursor (Tumor endothelial marker 7) (Tumor endothelial marker 3). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.006128 (rank : 154) | θ value | 0.47712 (rank : 145) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
TAF9_MOUSE
|
||||||
| NC score | 0.005814 (rank : 155) | θ value | 4.03905 (rank : 156) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VI33, Q32P09, Q80XS3, Q9CV61 | Gene names | Taf9, Taf2g | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor TFIID subunit 9 (Transcription initiation factor TFIID 31 kDa subunit) (TAFII-31) (TAFII-32) (TAFII32). | |||||
|
FA29A_HUMAN
|
||||||
| NC score | 0.005624 (rank : 156) | θ value | 6.88961 (rank : 160) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z4H7, Q6IQ10, Q6NZX5, Q8IZQ4, Q96FN0, Q9H950, Q9H998, Q9HCJ8, Q9NXT8 | Gene names | FAM29A, KIAA1574 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM29A. | |||||
|
ZYX_MOUSE
|
||||||
| NC score | 0.004189 (rank : 157) | θ value | 8.99809 (rank : 174) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62523, P70461 | Gene names | Zyx | |||
|
Domain Architecture |

|
|||||
| Description | Zyxin. | |||||
|
SON_MOUSE
|
||||||
| NC score | 0.003898 (rank : 158) | θ value | 1.06291 (rank : 146) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 957 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9QX47, Q9CQ12, Q9CQK6, Q9QXP5 | Gene names | Son | |||
|
Domain Architecture |

|
|||||
| Description | SON protein. | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.003886 (rank : 159) | θ value | 4.03905 (rank : 155) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
CAN2_HUMAN
|
||||||
| NC score | 0.003333 (rank : 160) | θ value | 4.03905 (rank : 152) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17655, Q16738, Q6PJT3, Q8WU26, Q9HBB1 | Gene names | CAPN2, CANPL2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (Calpain large polypeptide L2). | |||||
|
CAN2_MOUSE
|
||||||
| NC score | 0.002510 (rank : 161) | θ value | 6.88961 (rank : 159) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08529, O35518, O54843 | Gene names | Capn2 | |||
|
Domain Architecture |

|
|||||
| Description | Calpain-2 catalytic subunit precursor (EC 3.4.22.53) (Calpain-2 large subunit) (Calcium-activated neutral proteinase 2) (CANP 2) (Calpain M- type) (M-calpain) (Millimolar-calpain) (80 kDa M-calpain subunit) (CALP80). | |||||
|
MAGI1_MOUSE
|
||||||
| NC score | 0.002266 (rank : 162) | θ value | 0.21417 (rank : 143) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6RHR9, O54893, O54894, O54895 | Gene names | Magi1, Baiap1, Bap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (BAI1-associated protein 1) (BAP-1) (Membrane-associated guanylate kinase inverted 1) (MAGI-1). | |||||
|
ZN452_HUMAN
|
||||||
| NC score | 0.001242 (rank : 163) | θ value | 2.36792 (rank : 150) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6R2W3, Q2NKL9, Q5SRJ3, Q8TCN2, Q96PW3 | Gene names | ZNF452, KIAA1925 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF452. | |||||
|
MAML3_HUMAN
|
||||||
| NC score | 0.000690 (rank : 164) | θ value | 8.99809 (rank : 170) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96JK9 | Gene names | MAML3, KIAA1816 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mastermind-like protein 3 (Mam-3). | |||||
|
TS101_HUMAN
|
||||||
| NC score | 0.000414 (rank : 165) | θ value | 6.88961 (rank : 162) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99816, Q9BUM5 | Gene names | TSG101 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor susceptibility gene 101 protein. | |||||
|
SMRC2_HUMAN
|
||||||
| NC score | 0.000070 (rank : 166) | θ value | 5.27518 (rank : 158) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 499 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TAQ2, Q92923, Q96E12, Q96GY4 | Gene names | SMARCC2, BAF170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 2 (SWI/SNF complex 170 kDa subunit) (BRG1-associated factor 170). | |||||
|
MARCO_MOUSE
|
||||||
| NC score | -0.000124 (rank : 167) | θ value | 1.81305 (rank : 148) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q60754 | Gene names | Marco | |||
|
Domain Architecture |

|
|||||
| Description | Macrophage receptor MARCO (Macrophage receptor with collagenous structure). | |||||
|
NR4A3_HUMAN
|
||||||
| NC score | -0.000972 (rank : 168) | θ value | 4.03905 (rank : 154) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q92570, Q12935, Q14979, Q16420, Q4VXA9, Q9UEK2, Q9UEK3 | Gene names | NR4A3, CHN, CSMF, MINOR, NOR1, TEC | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A3 (Nuclear hormone receptor NOR-1) (Neuron-derived orphan receptor 1) (Mitogen-induced nuclear orphan receptor). | |||||
|
CO5A3_HUMAN
|
||||||
| NC score | -0.001094 (rank : 169) | θ value | 8.99809 (rank : 165) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 433 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P25940, Q9NZQ6 | Gene names | COL5A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(V) chain precursor. | |||||
|
CO4A2_MOUSE
|
||||||
| NC score | -0.001384 (rank : 170) | θ value | 5.27518 (rank : 157) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 492 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P08122, Q61375 | Gene names | Col4a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(IV) chain precursor. | |||||
|
NR4A3_MOUSE
|
||||||
| NC score | -0.001514 (rank : 171) | θ value | 8.99809 (rank : 171) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9QZB6, Q9QZB5 | Gene names | Nr4a3, Tec | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR4A3 (Orphan nuclear receptor TEC) (Translocated in extraskeletal chondrosarcoma). | |||||
|
CO9A3_HUMAN
|
||||||
| NC score | -0.001798 (rank : 172) | θ value | 8.99809 (rank : 166) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
IRX2_HUMAN
|
||||||
| NC score | -0.001930 (rank : 173) | θ value | 8.99809 (rank : 168) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BZI1, Q7Z2I7 | Gene names | IRX2, IRXA2 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-2 (Iroquois homeobox protein 2) (Homeodomain protein IRXA2). | |||||
|
ATS14_HUMAN
|
||||||
| NC score | -0.002311 (rank : 174) | θ value | 8.99809 (rank : 164) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8WXS8, Q8TE55, Q8TEY8 | Gene names | ADAMTS14 | |||
|
Domain Architecture |

|
|||||
| Description | ADAMTS-14 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase with thrombospondin motifs 14) (ADAM-TS 14) (ADAM-TS14). | |||||
|
SN_MOUSE
|
||||||
| NC score | -0.002873 (rank : 175) | θ value | 8.99809 (rank : 172) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 416 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62230, O55216, Q62228, Q62229 | Gene names | Sn, Sa | |||
|
Domain Architecture |

|
|||||
| Description | Sialoadhesin precursor (Sialic acid-binding Ig-like lectin-1) (Siglec- 1) (Sheep erythrocyte receptor) (SER) (CD169 antigen). | |||||
|
MINT_HUMAN
|
||||||
| NC score | -0.003314 (rank : 176) | θ value | 6.88961 (rank : 161) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1291 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96T58, Q9H9A8, Q9NWH5, Q9UQ01, Q9Y556 | Gene names | SPEN, KIAA0929, MINT, SHARP | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
KRHB6_MOUSE
|
||||||
| NC score | -0.004206 (rank : 177) | θ value | 8.99809 (rank : 169) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P97861, Q3TTV9 | Gene names | Krthb6, Krt2-10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cuticular Hb6 (Hair keratin, type II Hb6). | |||||
|
ZN446_HUMAN
|
||||||
| NC score | -0.008697 (rank : 178) | θ value | 8.99809 (rank : 173) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NWS9 | Gene names | ZNF446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 446. | |||||
|
EGR2_HUMAN
|
||||||
| NC score | -0.009890 (rank : 179) | θ value | 8.99809 (rank : 167) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 728 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11161, Q8IV26, Q9UNA6 | Gene names | EGR2, KROX20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early growth response protein 2 (EGR-2) (Krox-20 protein) (AT591). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | -0.010549 (rank : 180) | θ value | 8.99809 (rank : 163) | |||
| Query Neighborhood Hits | 174 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||