Please be patient as the page loads
|
DPOD3_MOUSE
|
||||||
| SwissProt Accessions | Q9EQ28 | Gene names | Pold3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DPOD3_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q9EQ28 | Gene names | Pold3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
DPOD3_HUMAN
|
||||||
| θ value | 3.47146e-177 (rank : 2) | NC score | 0.931224 (rank : 2) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15054 | Gene names | POLD3, KIAA0039 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | 5.44631e-05 (rank : 3) | NC score | 0.152360 (rank : 4) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
VEZA_HUMAN
|
||||||
| θ value | 0.00228821 (rank : 4) | NC score | 0.143317 (rank : 5) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HBM0, Q9H2F4, Q9H2U5, Q9NT70, Q9NVW0, Q9UF91 | Gene names | VEZT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vezatin. | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 5) | NC score | 0.153804 (rank : 3) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 6) | NC score | 0.085570 (rank : 32) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
H12_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 7) | NC score | 0.100320 (rank : 17) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P15864 | Gene names | Hist1h1c, H1f2 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.2 (H1 VAR.1) (H1c). | |||||
|
MAP2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 8) | NC score | 0.079359 (rank : 40) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
TXND2_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 9) | NC score | 0.111813 (rank : 13) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
H13_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 10) | NC score | 0.103507 (rank : 16) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
PRR12_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 11) | NC score | 0.095520 (rank : 22) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
CJ012_HUMAN
|
||||||
| θ value | 0.125558 (rank : 12) | NC score | 0.124853 (rank : 6) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 13) | NC score | 0.114242 (rank : 11) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 0.163984 (rank : 14) | NC score | 0.119242 (rank : 10) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
H12_HUMAN
|
||||||
| θ value | 0.21417 (rank : 15) | NC score | 0.094061 (rank : 23) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P16403 | Gene names | HIST1H1C, H1F2 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.2 (Histone H1d). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | 0.21417 (rank : 16) | NC score | 0.096523 (rank : 20) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
ATX2L_HUMAN
|
||||||
| θ value | 0.279714 (rank : 17) | NC score | 0.071425 (rank : 54) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
CAF1A_HUMAN
|
||||||
| θ value | 0.279714 (rank : 18) | NC score | 0.072239 (rank : 53) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
MDC1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 19) | NC score | 0.084326 (rank : 34) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.100093 (rank : 18) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
ANK3_HUMAN
|
||||||
| θ value | 0.365318 (rank : 21) | NC score | 0.031185 (rank : 143) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
HRX_HUMAN
|
||||||
| θ value | 0.365318 (rank : 22) | NC score | 0.075680 (rank : 47) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 0.365318 (rank : 23) | NC score | 0.078576 (rank : 42) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.042884 (rank : 134) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.111209 (rank : 14) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 0.47712 (rank : 26) | NC score | 0.052852 (rank : 106) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
MYO9B_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.036999 (rank : 138) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
NOP56_MOUSE
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.060455 (rank : 80) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
BASP_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.121482 (rank : 8) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
PHF14_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.048440 (rank : 127) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
RD23B_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.049484 (rank : 126) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
RFC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.083589 (rank : 35) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.013693 (rank : 176) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 34) | NC score | 0.096391 (rank : 21) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
UBIP1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 35) | NC score | 0.047798 (rank : 128) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZI7, Q68CT0, Q86Y57, Q9H8V0, Q9UD76, Q9UD78 | Gene names | UBP1, LBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (LBP-1). | |||||
|
UBIP1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 36) | NC score | 0.041920 (rank : 135) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q811S7, Q3UPR3, Q3US11, Q60786, Q8C514 | Gene names | Ubp1, Cp2b, Nf2d9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (Nuclear factor 2d9) (NF2d9). | |||||
|
DDX21_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.033013 (rank : 141) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.073984 (rank : 51) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
ZN75A_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.001453 (rank : 187) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96N20, Q92669 | Gene names | ZNF75A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 75A. | |||||
|
ENL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.113718 (rank : 12) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
H13_HUMAN
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.086758 (rank : 26) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P16402 | Gene names | HIST1H1D, H1F3 | |||
|
Domain Architecture |
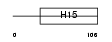
|
|||||
| Description | Histone H1.3 (Histone H1c). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 42) | NC score | 0.104764 (rank : 15) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 43) | NC score | 0.121610 (rank : 7) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.082108 (rank : 37) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
FMNL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 45) | NC score | 0.025946 (rank : 154) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
MOR2A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 46) | NC score | 0.028827 (rank : 148) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q69ZX6, Q5QNQ7, Q6P547 | Gene names | Morc2a, Kiaa0852, Zcwcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2A (Zinc finger CW-type coiled-coil domain protein 1). | |||||
|
MPP8_HUMAN
|
||||||
| θ value | 1.81305 (rank : 47) | NC score | 0.045879 (rank : 131) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 48) | NC score | 0.086183 (rank : 29) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
AL2S4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.062330 (rank : 74) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
ATX2L_MOUSE
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.056010 (rank : 90) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
CE152_HUMAN
|
||||||
| θ value | 2.36792 (rank : 51) | NC score | 0.025355 (rank : 156) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
MRP_MOUSE
|
||||||
| θ value | 2.36792 (rank : 52) | NC score | 0.061846 (rank : 75) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
NEBU_HUMAN
|
||||||
| θ value | 2.36792 (rank : 53) | NC score | 0.025540 (rank : 155) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P20929, Q15346 | Gene names | NEB | |||
|
Domain Architecture |
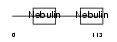
|
|||||
| Description | Nebulin. | |||||
|
PREB_MOUSE
|
||||||
| θ value | 2.36792 (rank : 54) | NC score | 0.029159 (rank : 146) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUQ2, Q9QZ99 | Gene names | Preb, Sec12 | |||
|
Domain Architecture |
|
|||||
| Description | Prolactin regulatory element-binding protein (Mammalian guanine nucleotide exchange factor mSec12). | |||||
|
ROCK2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 55) | NC score | 0.013407 (rank : 177) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
ATE1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 56) | NC score | 0.050807 (rank : 119) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2A5, Q9Z2A4 | Gene names | Ate1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl-tRNA--protein transferase 1 (EC 2.3.2.8) (R-transferase 1) (Arginyltransferase 1) (Arginine-tRNA--protein transferase 1). | |||||
|
CHD6_HUMAN
|
||||||
| θ value | 3.0926 (rank : 57) | NC score | 0.026642 (rank : 151) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
DNL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 58) | NC score | 0.074043 (rank : 50) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
HXA13_MOUSE
|
||||||
| θ value | 3.0926 (rank : 59) | NC score | 0.012142 (rank : 180) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62424 | Gene names | Hoxa13, Hox-1.10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A13 (Hox-1.10). | |||||
|
MAGD2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 60) | NC score | 0.018457 (rank : 169) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
MARCS_HUMAN
|
||||||
| θ value | 3.0926 (rank : 61) | NC score | 0.081699 (rank : 38) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MUG1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 62) | NC score | 0.016117 (rank : 173) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28665 | Gene names | Mug1, Mug-1 | |||
|
Domain Architecture |

|
|||||
| Description | Murinoglobulin-1 precursor (MuG1). | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 63) | NC score | 0.028270 (rank : 149) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 64) | NC score | 0.044910 (rank : 133) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
SASH1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 65) | NC score | 0.031998 (rank : 142) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O94885, Q8TAI0, Q9H7R7 | Gene names | SASH1, KIAA0790, PEPE1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1 (Proline-glutamate repeat- containing protein). | |||||
|
TDG_HUMAN
|
||||||
| θ value | 3.0926 (rank : 66) | NC score | 0.046842 (rank : 129) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13569, Q8IUZ6, Q8IZM3 | Gene names | TDG | |||
|
Domain Architecture |
|
|||||
| Description | G/T mismatch-specific thymine DNA glycosylase (EC 3.2.2.-). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | 3.0926 (rank : 67) | NC score | 0.088899 (rank : 25) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
BRD2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 68) | NC score | 0.030245 (rank : 145) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 69) | NC score | 0.050634 (rank : 120) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CAR10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 70) | NC score | 0.034473 (rank : 140) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BWT7, Q14CQ8, Q5TFG6, Q9UGR5, Q9UGR6, Q9Y3H0 | Gene names | CARD10, CARMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (CARD-containing MAGUK protein 3) (Carma 3). | |||||
|
KI67_HUMAN
|
||||||
| θ value | 4.03905 (rank : 71) | NC score | 0.119717 (rank : 9) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 72) | NC score | 0.086032 (rank : 30) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NFH_HUMAN
|
||||||
| θ value | 4.03905 (rank : 73) | NC score | 0.064475 (rank : 67) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 4.03905 (rank : 74) | NC score | 0.046548 (rank : 130) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
PHF22_HUMAN
|
||||||
| θ value | 4.03905 (rank : 75) | NC score | 0.035524 (rank : 139) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96CB8, Q9HD71 | Gene names | INTS12, PHF22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
RFC1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 76) | NC score | 0.062566 (rank : 72) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
RRP5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 77) | NC score | 0.056641 (rank : 87) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
RYR3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 78) | NC score | 0.017073 (rank : 172) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
SPEG_MOUSE
|
||||||
| θ value | 4.03905 (rank : 79) | NC score | 0.018027 (rank : 170) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
ZO2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 80) | NC score | 0.019395 (rank : 166) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
ANK2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 81) | NC score | 0.050291 (rank : 125) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 82) | NC score | 0.019169 (rank : 167) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 83) | NC score | 0.022136 (rank : 160) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CENPO_MOUSE
|
||||||
| θ value | 5.27518 (rank : 84) | NC score | 0.026477 (rank : 152) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K015, Q8R587, Q9CYY6 | Gene names | Cenpo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein O (CENP-O). | |||||
|
CJ118_HUMAN
|
||||||
| θ value | 5.27518 (rank : 85) | NC score | 0.017239 (rank : 171) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
CTRO_HUMAN
|
||||||
| θ value | 5.27518 (rank : 86) | NC score | 0.010357 (rank : 182) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
FGD6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 87) | NC score | 0.014803 (rank : 174) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 5.27518 (rank : 88) | NC score | 0.086250 (rank : 28) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
MCF2L_MOUSE
|
||||||
| θ value | 5.27518 (rank : 89) | NC score | 0.013977 (rank : 175) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64096 | Gene names | Mcf2l, Dbs | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide exchange factor DBS (DBL's big sister) (MCF2- transforming sequence-like protein). | |||||
|
NUCL_MOUSE
|
||||||
| θ value | 5.27518 (rank : 90) | NC score | 0.026047 (rank : 153) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
PHF20_MOUSE
|
||||||
| θ value | 5.27518 (rank : 91) | NC score | 0.075356 (rank : 48) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
PKCB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 92) | NC score | 0.052049 (rank : 108) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 93) | NC score | 0.076370 (rank : 45) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
AFF2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 94) | NC score | 0.045198 (rank : 132) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
CCD27_MOUSE
|
||||||
| θ value | 6.88961 (rank : 95) | NC score | 0.019583 (rank : 165) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
HXA13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 96) | NC score | 0.010311 (rank : 183) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31271, O43371 | Gene names | HOXA13, HOX1J | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A13 (Hox-1J). | |||||
|
PCD19_HUMAN
|
||||||
| θ value | 6.88961 (rank : 97) | NC score | 0.004121 (rank : 185) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAB3, Q5JTG1, Q5JTG2, Q68DT7, Q9P2N3 | Gene names | PCDH19, KIAA1313 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-19 precursor. | |||||
|
PKHA6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 98) | NC score | 0.018627 (rank : 168) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
RENT2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 99) | NC score | 0.041724 (rank : 136) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
ABCF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 100) | NC score | 0.029080 (rank : 147) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 101) | NC score | 0.040042 (rank : 137) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
BRE1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 102) | NC score | 0.020776 (rank : 162) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
CAR10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 103) | NC score | 0.021957 (rank : 161) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P58660 | Gene names | Card10, Bimp1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (Bcl10-interacting MAGUK protein 1) (Bimp1). | |||||
|
CCD21_HUMAN
|
||||||
| θ value | 8.99809 (rank : 104) | NC score | 0.020603 (rank : 164) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CT075_MOUSE
|
||||||
| θ value | 8.99809 (rank : 105) | NC score | 0.007085 (rank : 184) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59383 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf75 homolog precursor. | |||||
|
CTCF_HUMAN
|
||||||
| θ value | 8.99809 (rank : 106) | NC score | -0.000050 (rank : 188) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49711 | Gene names | CTCF | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||
|
HMGB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 107) | NC score | 0.012345 (rank : 179) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26583 | Gene names | HMGB2, HMG2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HMGB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 108) | NC score | 0.011303 (rank : 181) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30681, Q3UXT1, Q9EQD5 | Gene names | Hmgb2, Hmg2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HUCE1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 109) | NC score | 0.054745 (rank : 97) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DB85, Q8BHW3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebral protein 1 homolog. | |||||
|
MDC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 110) | NC score | 0.080226 (rank : 39) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
NCOR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 111) | NC score | 0.026979 (rank : 150) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
NO40_MOUSE
|
||||||
| θ value | 8.99809 (rank : 112) | NC score | 0.023953 (rank : 158) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ESX4 | Gene names | Zcchc17, Ps1d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein of 40 kDa (pNO40) (Zinc finger CCHC domain- containing protein 17) (Putative S1 RNA-binding domain protein) (PS1D protein). | |||||
|
PCD19_MOUSE
|
||||||
| θ value | 8.99809 (rank : 113) | NC score | 0.003929 (rank : 186) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80TF3, Q8BL13 | Gene names | Pcdh19, Kiaa1313 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-19 precursor. | |||||
|
PLCB1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 114) | NC score | 0.022968 (rank : 159) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
PLCB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 115) | NC score | 0.020720 (rank : 163) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
PNRC1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 116) | NC score | 0.025256 (rank : 157) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12796, Q5T7J6 | Gene names | PNRC1, PROL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich nuclear receptor coactivator 1 (Proline-rich protein 2) (B4-2 protein). | |||||
|
RGS3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 117) | NC score | 0.030839 (rank : 144) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 118) | NC score | 0.013367 (rank : 178) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.059788 (rank : 84) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
AF9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.064015 (rank : 68) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
AFF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.062580 (rank : 71) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
AFF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.051347 (rank : 112) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.073378 (rank : 52) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.068378 (rank : 57) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.078322 (rank : 43) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CF010_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.071087 (rank : 55) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.055374 (rank : 94) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CN155_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.055451 (rank : 92) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.055430 (rank : 93) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.067001 (rank : 61) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DEK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.051406 (rank : 111) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
DEK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.051232 (rank : 114) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
DMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.051039 (rank : 117) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.061322 (rank : 77) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.050421 (rank : 124) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
H10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.061648 (rank : 76) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P07305 | Gene names | H1F0, H1FV | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.0 (H1(0)) (Histone H1'). | |||||
|
H10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.062345 (rank : 73) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10922 | Gene names | H1f0, H1fv | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1' (H1.0) (H1(0)). | |||||
|
H11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.067806 (rank : 60) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02539 | Gene names | HIST1H1A, H1F1 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.1. | |||||
|
H11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.064975 (rank : 65) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P43275 | Gene names | Hist1h1a, H1f1 | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1.1 (H1 VAR.3) (H1a). | |||||
|
H14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.075941 (rank : 46) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
H14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.076726 (rank : 44) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
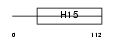
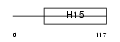
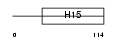
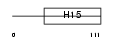
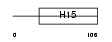
H15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.078876 (rank : 41) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
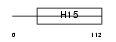
H15_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.086385 (rank : 27) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
H1T_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.051762 (rank : 110) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22492, Q8IUE8 | Gene names | HIST1H1T, H1FT, H1T | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1t (Testicular H1 histone). | |||||
|
H1T_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.051137 (rank : 116) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q07133, Q8CGP8 | Gene names | Hist1h1t, H1ft, H1t | |||
|
Domain Architecture |
|
|||||
| Description | Histone H1t (Testicular H1 histone). | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.061257 (rank : 78) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
ICAL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.053022 (rank : 105) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
K1210_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.056636 (rank : 88) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
LAD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.084752 (rank : 33) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.055496 (rank : 91) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MAP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.050951 (rank : 118) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
MAP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.053389 (rank : 102) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.053126 (rank : 104) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
MBB1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.054796 (rank : 95) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
MINT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.051279 (rank : 113) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
MRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.052044 (rank : 109) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
NASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.066979 (rank : 62) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.059961 (rank : 83) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.054768 (rank : 96) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.085864 (rank : 31) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.054736 (rank : 98) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.068263 (rank : 58) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.060350 (rank : 82) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
PAF49_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.053738 (rank : 100) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
PRG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.052316 (rank : 107) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PRG4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.053903 (rank : 99) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.092768 (rank : 24) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RL1D1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.051203 (rank : 115) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.059175 (rank : 85) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SET1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.057975 (rank : 86) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
SFR11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.060360 (rank : 81) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.056239 (rank : 89) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SFR17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.050558 (rank : 123) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.065126 (rank : 64) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
SRRM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.063200 (rank : 70) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.063707 (rank : 69) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
T2FA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.060659 (rank : 79) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
TARA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.053346 (rank : 103) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
TAU_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.053691 (rank : 101) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.083586 (rank : 36) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TGON1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.074293 (rank : 49) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
TGON2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.068716 (rank : 56) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
THOC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.050624 (rank : 121) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
TXND2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.098959 (rank : 19) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.065593 (rank : 63) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
VCX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.064953 (rank : 66) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
VCXC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.068244 (rank : 59) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.050581 (rank : 122) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
DPOD3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q9EQ28 | Gene names | Pold3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
DPOD3_HUMAN
|
||||||
| NC score | 0.931224 (rank : 2) | θ value | 3.47146e-177 (rank : 2) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 109 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15054 | Gene names | POLD3, KIAA0039 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA polymerase subunit delta 3 (DNA polymerase subunit delta p66). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.153804 (rank : 3) | θ value | 0.0330416 (rank : 5) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.152360 (rank : 4) | θ value | 5.44631e-05 (rank : 3) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
VEZA_HUMAN
|
||||||
| NC score | 0.143317 (rank : 5) | θ value | 0.00228821 (rank : 4) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HBM0, Q9H2F4, Q9H2U5, Q9NT70, Q9NVW0, Q9UF91 | Gene names | VEZT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vezatin. | |||||
|
CJ012_HUMAN
|
||||||
| NC score | 0.124853 (rank : 6) | θ value | 0.125558 (rank : 12) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q8N655, Q9H945, Q9Y457 | Gene names | C10orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf12. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.121610 (rank : 7) | θ value | 1.38821 (rank : 43) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.121482 (rank : 8) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
KI67_HUMAN
|
||||||
| NC score | 0.119717 (rank : 9) | θ value | 4.03905 (rank : 71) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P46013 | Gene names | MKI67 | |||
|
Domain Architecture |

|
|||||
| Description | Antigen KI-67. | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.119242 (rank : 10) | θ value | 0.163984 (rank : 14) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.114242 (rank : 11) | θ value | 0.163984 (rank : 13) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.113718 (rank : 12) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.111813 (rank : 13) | θ value | 0.0736092 (rank : 9) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.111209 (rank : 14) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.104764 (rank : 15) | θ value | 1.38821 (rank : 42) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
H13_MOUSE
|
||||||
| NC score | 0.103507 (rank : 16) | θ value | 0.0961366 (rank : 10) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
H12_MOUSE
|
||||||
| NC score | 0.100320 (rank : 17) | θ value | 0.0736092 (rank : 7) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 52 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P15864 | Gene names | Hist1h1c, H1f2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.2 (H1 VAR.1) (H1c). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.100093 (rank : 18) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
TXND2_HUMAN
|
||||||
| NC score | 0.098959 (rank : 19) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 793 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q86VQ3, Q8N7U4, Q96RX3, Q9H0L8 | Gene names | TXNDC2, SPTRX, SPTRX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.096523 (rank : 20) | θ value | 0.21417 (rank : 16) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.096391 (rank : 21) | θ value | 0.813845 (rank : 34) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
PRR12_HUMAN
|
||||||
| NC score | 0.095520 (rank : 22) | θ value | 0.0961366 (rank : 11) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 460 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ULL5, Q8N4J6 | Gene names | PRR12, KIAA1205 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 12. | |||||
|
H12_HUMAN
|
||||||
| NC score | 0.094061 (rank : 23) | θ value | 0.21417 (rank : 15) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P16403 | Gene names | HIST1H1C, H1F2 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.2 (Histone H1d). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.092768 (rank : 24) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.088899 (rank : 25) | θ value | 3.0926 (rank : 67) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
H13_HUMAN
|
||||||
| NC score | 0.086758 (rank : 26) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P16402 | Gene names | HIST1H1D, H1F3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (Histone H1c). | |||||
|
H15_MOUSE
|
||||||
| NC score | 0.086385 (rank : 27) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P43276, Q9CRM8 | Gene names | Hist1h1b, H1f5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (H1 VAR.5) (H1b). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.086250 (rank : 28) | θ value | 5.27518 (rank : 88) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.086183 (rank : 29) | θ value | 1.81305 (rank : 48) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.086032 (rank : 30) | θ value | 4.03905 (rank : 72) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.085864 (rank : 31) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.085570 (rank : 32) | θ value | 0.0563607 (rank : 6) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.084752 (rank : 33) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
MDC1_MOUSE
|
||||||
| NC score | 0.084326 (rank : 34) | θ value | 0.279714 (rank : 19) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 998 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q5PSV9, Q5U4D3, Q6ZQH7 | Gene names | Mdc1, Kiaa0170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1. | |||||
|
RFC1_HUMAN
|
||||||
| NC score | 0.083589 (rank : 35) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 352 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P35251, Q5XKF5, Q6PKU0, Q86V41, Q86V46 | Gene names | RFC1, RFC140 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (DNA-binding protein PO-GA). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.083586 (rank : 36) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.082108 (rank : 37) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.081699 (rank : 38) | θ value | 3.0926 (rank : 61) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
MDC1_HUMAN
|
||||||
| NC score | 0.080226 (rank : 39) | θ value | 8.99809 (rank : 110) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1446 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q14676, Q5JP55, Q5JP56, Q5ST83, Q68CQ3, Q86Z06, Q96QC2 | Gene names | MDC1, KIAA0170, NFBD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mediator of DNA damage checkpoint protein 1 (Nuclear factor with BRCT domains 1). | |||||
|
MAP2_MOUSE
|
||||||
| NC score | 0.079359 (rank : 40) | θ value | 0.0736092 (rank : 8) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
H15_HUMAN
|
||||||
| NC score | 0.078876 (rank : 41) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16401, Q14529 | Gene names | HIST1H1B, H1F5 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.5 (Histone H1a). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.078576 (rank : 42) | θ value | 0.365318 (rank : 23) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.078322 (rank : 43) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
H14_MOUSE
|
||||||
| NC score | 0.076726 (rank : 44) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P43274 | Gene names | Hist1h1e, H1f4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (H1 VAR.2) (H1e). | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.076370 (rank : 45) | θ value | 5.27518 (rank : 93) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
H14_HUMAN
|
||||||
| NC score | 0.075941 (rank : 46) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
HRX_HUMAN
|
||||||
| NC score | 0.075680 (rank : 47) | θ value | 0.365318 (rank : 22) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q03164, Q13743, Q13744, Q14845, Q16364, Q6UBD1, Q9UMA3 | Gene names | MLL, ALL1, HRX, HTRX, TRX1 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein HRX (ALL-1) (Trithorax-like protein). | |||||
|
PHF20_MOUSE
|
||||||
| NC score | 0.075356 (rank : 48) | θ value | 5.27518 (rank : 91) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 513 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q8BLG0, Q8BMA2, Q8BYR4, Q921N1 | Gene names | Phf20, Hca58 | |||
|
Domain Architecture |

|
|||||
| Description | PHD finger protein 20 (Hepatocellular carcinoma-associated antigen 58 homolog). | |||||
|
TGON1_MOUSE
|
||||||
| NC score | 0.074293 (rank : 49) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q62313 | Gene names | Tgoln1, Ttgn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 1 precursor (TGN38A). | |||||
|
DNL1_HUMAN
|
||||||
| NC score | 0.074043 (rank : 50) | θ value | 3.0926 (rank : 58) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P18858 | Gene names | LIG1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA ligase 1 (EC 6.5.1.1) (DNA ligase I) (Polydeoxyribonucleotide synthase [ATP] 1). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.073984 (rank : 51) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.073378 (rank : 52) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
CAF1A_HUMAN
|
||||||
| NC score | 0.072239 (rank : 53) | θ value | 0.279714 (rank : 18) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 750 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q13111, Q7Z7K3, Q9UJY8 | Gene names | CHAF1A, CAF, CAF1P150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
ATX2L_HUMAN
|
||||||
| NC score | 0.071425 (rank : 54) | θ value | 0.279714 (rank : 17) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 443 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WWM7, O95135, Q6NVJ8, Q6PJW6, Q8IU61, Q8IU95, Q8WWM3, Q8WWM4, Q8WWM5, Q8WWM6, Q99703 | Gene names | ATXN2L, A2D, A2LG, A2LP, A2RP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein (Ataxin-2 domain protein) (Ataxin-2-related protein). | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.071087 (rank : 55) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
TGON2_MOUSE
|
||||||
| NC score | 0.068716 (rank : 56) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 277 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q62314 | Gene names | Tgoln2, Ttgn2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (TGN38B). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.068378 (rank : 57) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.068263 (rank : 58) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
VCXC_HUMAN
|
||||||
| NC score | 0.068244 (rank : 59) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
H11_HUMAN
|
||||||
| NC score | 0.067806 (rank : 60) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q02539 | Gene names | HIST1H1A, H1F1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.1. | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.067001 (rank : 61) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.066979 (rank : 62) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.065593 (rank : 63) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.065126 (rank : 64) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
H11_MOUSE
|
||||||
| NC score | 0.064975 (rank : 65) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P43275 | Gene names | Hist1h1a, H1f1 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.1 (H1 VAR.3) (H1a). | |||||
|
VCX3_HUMAN
|
||||||
| NC score | 0.064953 (rank : 66) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.064475 (rank : 67) | θ value | 4.03905 (rank : 73) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
AF9_HUMAN
|
||||||
| NC score | 0.064015 (rank : 68) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P42568 | Gene names | MLLT3, AF9, YEATS3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein AF-9 (ALL1 fused gene from chromosome 9 protein) (Myeloid/lymphoid or mixed-lineage leukemia translocated to chromosome 3 protein) (YEATS domain-containing protein 3). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.063707 (rank : 69) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
SRRM1_HUMAN
|
||||||
| NC score | 0.063200 (rank : 70) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8IYB3, O60585, Q5VVN4 | Gene names | SRRM1, SRM160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Ser/Arg-related nuclear matrix protein) (SR-related nuclear matrix protein of 160 kDa) (SRm160). | |||||
|
AFF1_HUMAN
|
||||||
| NC score | 0.062580 (rank : 71) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 506 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P51825 | Gene names | AFF1, AF4, FEL, MLLT2 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4) (Protein FEL). | |||||
|
RFC1_MOUSE
|
||||||
| NC score | 0.062566 (rank : 72) | θ value | 4.03905 (rank : 76) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P35601 | Gene names | Rfc1, Ibf-1, Recc1 | |||
|
Domain Architecture |

|
|||||
| Description | Replication factor C subunit 1 (Replication factor C large subunit) (RF-C 140 kDa subunit) (Activator 1 140 kDa subunit) (Activator 1 large subunit) (A1 140 kDa subunit) (A1-P145) (Differentiation- specific element-binding protein) (ISRE-binding protein). | |||||
|
H10_MOUSE
|
||||||
| NC score | 0.062345 (rank : 73) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 50 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P10922 | Gene names | H1f0, H1fv | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1' (H1.0) (H1(0)). | |||||
|
AL2S4_MOUSE
|
||||||
| NC score | 0.062330 (rank : 74) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q3V0J1, Q3TIS2 | Gene names | Als2cr4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyotrophic lateral sclerosis 2 chromosomal region candidate gene 4 protein homolog. | |||||
|
MRP_MOUSE
|
||||||
| NC score | 0.061846 (rank : 75) | θ value | 2.36792 (rank : 52) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
H10_HUMAN
|
||||||
| NC score | 0.061648 (rank : 76) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P07305 | Gene names | H1F0, H1FV | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.0 (H1(0)) (Histone H1'). | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.061322 (rank : 77) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.061257 (rank : 78) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.060659 (rank : 79) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
NOP56_MOUSE
|
||||||
| NC score | 0.060455 (rank : 80) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.060360 (rank : 81) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.060350 (rank : 82) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.059961 (rank : 83) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.059788 (rank : 84) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.059175 (rank : 85) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SET1A_HUMAN
|
||||||
| NC score | 0.057975 (rank : 86) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 690 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O15047, Q6PIF3, Q8TAJ6 | Gene names | SETD1A, KIAA0339, SET1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-4 specific SET1 (EC 2.1.1.43) (Set1/Ash2 histone methyltransferase complex subunit SET1) (SET-domain-containing protein 1A). | |||||
|
RRP5_HUMAN
|
||||||
| NC score | 0.056641 (rank : 87) | θ value | 4.03905 (rank : 77) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q14690, Q86VQ8 | Gene names | PDCD11, KIAA0185 | |||
|
Domain Architecture |

|
|||||
| Description | RRP5 protein homolog (Programmed cell death protein 11). | |||||
|
K1210_HUMAN
|
||||||
| NC score | 0.056636 (rank : 88) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9ULL0, Q5JPN4 | Gene names | KIAA1210 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA1210. | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.056239 (rank : 89) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
ATX2L_MOUSE
|
||||||
| NC score | 0.056010 (rank : 90) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7TQH0, Q80XN9, Q8K059 | Gene names | Atxn2l, A2lp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ataxin-2-like protein. | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.055496 (rank : 91) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
CN155_HUMAN
|
||||||
| NC score | 0.055451 (rank : 92) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q5H9T9, Q5H9U7, Q86YI2, Q9H0J3 | Gene names | C14orf155 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf155. | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.055430 (rank : 93) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.055374 (rank : 94) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
MBB1A_HUMAN
|
||||||
| NC score | 0.054796 (rank : 95) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 665 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9BQG0, Q86VM3, Q9BW49, Q9P0V5, Q9UF99 | Gene names | MYBBP1A, P160 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myb-binding protein 1A. | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.054768 (rank : 96) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
HUCE1_MOUSE
|
||||||
| NC score | 0.054745 (rank : 97) | θ value | 8.99809 (rank : 109) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DB85, Q8BHW3 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cerebral protein 1 homolog. | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.054736 (rank : 98) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
PRG4_MOUSE
|
||||||
| NC score | 0.053903 (rank : 99) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q9JM99, Q3UEL1, Q3V198 | Gene names | Prg4, Msf, Szp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PAF49_HUMAN
|
||||||
| NC score | 0.053738 (rank : 100) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O15446, Q32N11, Q7Z5U2, Q9UPF6 | Gene names | CD3EAP, ASE1, CAST, PAF49 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase I-associated factor PAF49 (Anti-sense to ERCC-1 protein) (ASE-1) (CD3-epsilon-associated protein) (CD3E-associated protein) (CAST). | |||||
|
TAU_MOUSE
|
||||||
| NC score | 0.053691 (rank : 101) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 502 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P10637, P10638, Q60684, Q60685, Q60686, Q62286 | Gene names | Mapt, Mtapt, Tau | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein tau (Neurofibrillary tangle protein) (Paired helical filament-tau) (PHF-tau). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.053389 (rank : 102) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.053346 (rank : 103) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.053126 (rank : 104) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
ICAL_HUMAN
|
||||||
| NC score | 0.053022 (rank : 105) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P20810, O95360, Q96D08, Q9H1Z5 | Gene names | CAST | |||
|
Domain Architecture |

|
|||||
| Description | Calpastatin (Calpain inhibitor) (Sperm BS-17 component). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.052852 (rank : 106) | θ value | 0.47712 (rank : 26) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
PRG4_HUMAN
|
||||||
| NC score | 0.052316 (rank : 107) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92954, Q6DNC4, Q6DNC5, Q6ZMZ5, Q9BX49 | Gene names | PRG4, MSF, SZP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proteoglycan-4 precursor (Lubricin) (Megakaryocyte-stimulating factor) (Superficial zone proteoglycan) [Contains: Proteoglycan-4 C-terminal part]. | |||||
|
PKCB1_HUMAN
|
||||||
| NC score | 0.052049 (rank : 108) | θ value | 5.27518 (rank : 92) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 488 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9ULU4, Q13517, Q4JJ94, Q4JJ95, Q5TH09, Q6MZM1, Q8WXC5, Q9H1F3, Q9H1F4, Q9H1F5, Q9H1L8, Q9H1L9, Q9H2G5, Q9NYN3, Q9UIX6 | Gene names | PRKCBP1, KIAA1125, RACK7, ZMYND8 | |||
|
Domain Architecture |

|
|||||
| Description | Protein kinase C-binding protein 1 (Rack7) (Cutaneous T-cell lymphoma- associated antigen se14-3) (CTCL tumor antigen se14-3) (Zinc finger MYND domain-containing protein 8). | |||||
|
MRP_HUMAN
|
||||||
| NC score | 0.052044 (rank : 109) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
H1T_HUMAN
|
||||||
| NC score | 0.051762 (rank : 110) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P22492, Q8IUE8 | Gene names | HIST1H1T, H1FT, H1T | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1t (Testicular H1 histone). | |||||
|
DEK_HUMAN
|
||||||
| NC score | 0.051406 (rank : 111) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 199 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P35659, Q5TGV4, Q5TGV5 | Gene names | DEK | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
AFF1_MOUSE
|
||||||
| NC score | 0.051347 (rank : 112) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | O88573 | Gene names | Aff1, Mllt2, Mllt2h | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 1 (Protein AF-4) (Proto-oncogene AF4). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.051279 (rank : 113) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.051232 (rank : 114) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
RL1D1_HUMAN
|
||||||
| NC score | 0.051203 (rank : 115) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O76021, Q6PL22, Q8IWS7, Q8WUZ1, Q9HDA9, Q9Y3Z9 | Gene names | RSL1D1, CATX11, CSIG, PBK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ribosomal L1 domain-containing protein 1 (Cellular senescence- inhibited gene protein) (Protein PBK1) (CATX-11). | |||||
|
H1T_MOUSE
|
||||||
| NC score | 0.051137 (rank : 116) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q07133, Q8CGP8 | Gene names | Hist1h1t, H1ft, H1t | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1t (Testicular H1 histone). | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.051039 (rank : 117) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
MAP2_HUMAN
|
||||||
| NC score | 0.050951 (rank : 118) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P11137, Q99975, Q99976 | Gene names | MAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2) (MAP-2). | |||||
|
ATE1_MOUSE
|
||||||
| NC score | 0.050807 (rank : 119) | θ value | 3.0926 (rank : 56) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z2A5, Q9Z2A4 | Gene names | Ate1 | |||
|
Domain Architecture |

|
|||||
| Description | Arginyl-tRNA--protein transferase 1 (EC 2.3.2.8) (R-transferase 1) (Arginyltransferase 1) (Arginine-tRNA--protein transferase 1). | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.050634 (rank : 120) | θ value | 4.03905 (rank : 69) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.050624 (rank : 121) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.050581 (rank : 122) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
SFR17_HUMAN
|
||||||
| NC score | 0.050558 (rank : 123) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.050421 (rank : 124) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ANK2_HUMAN
|
||||||
| NC score | 0.050291 (rank : 125) | θ value | 5.27518 (rank : 81) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1615 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q01484, Q01485 | Gene names | ANK2 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-2 (Brain ankyrin) (Ankyrin-B) (Ankyrin, nonerythroid). | |||||
|
RD23B_MOUSE
|
||||||
| NC score | 0.049484 (rank : 126) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54728 | Gene names | Rad23b, Mhr23b | |||
|
Domain Architecture |

|
|||||
| Description | UV excision repair protein RAD23 homolog B (mHR23B) (XP-C repair- complementing complex 58 kDa protein) (p58). | |||||
|
PHF14_HUMAN
|
||||||
| NC score | 0.048440 (rank : 127) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O94880 | Gene names | PHF14, KIAA0783 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | PHD finger protein 14. | |||||
|
UBIP1_HUMAN
|
||||||
| NC score | 0.047798 (rank : 128) | θ value | 0.813845 (rank : 35) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NZI7, Q68CT0, Q86Y57, Q9H8V0, Q9UD76, Q9UD78 | Gene names | UBP1, LBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (LBP-1). | |||||
|
TDG_HUMAN
|
||||||
| NC score | 0.046842 (rank : 129) | θ value | 3.0926 (rank : 66) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q13569, Q8IUZ6, Q8IZM3 | Gene names | TDG | |||
|
Domain Architecture |

|
|||||
| Description | G/T mismatch-specific thymine DNA glycosylase (EC 3.2.2.-). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.046548 (rank : 130) | θ value | 4.03905 (rank : 74) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
MPP8_HUMAN
|
||||||
| NC score | 0.045879 (rank : 131) | θ value | 1.81305 (rank : 47) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 948 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q99549, Q86TK3, Q9BTP1 | Gene names | MPHOSPH8, MPP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | M-phase phosphoprotein 8. | |||||
|
AFF2_MOUSE
|
||||||
| NC score | 0.045198 (rank : 132) | θ value | 6.88961 (rank : 94) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O55112 | Gene names | Aff2, Fmr2, Ox19 | |||
|
Domain Architecture |

|
|||||
| Description | AF4/FMR2 family member 2 (Fragile X mental retardation protein 2 homolog) (Protein FMR-2) (FMR2P) (Protein Ox19). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.044910 (rank : 133) | θ value | 3.0926 (rank : 64) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.042884 (rank : 134) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
UBIP1_MOUSE
|
||||||
| NC score | 0.041920 (rank : 135) | θ value | 0.813845 (rank : 36) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q811S7, Q3UPR3, Q3US11, Q60786, Q8C514 | Gene names | Ubp1, Cp2b, Nf2d9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Upstream-binding protein 1 (Nuclear factor 2d9) (NF2d9). | |||||
|
RENT2_HUMAN
|
||||||
| NC score | 0.041724 (rank : 136) | θ value | 6.88961 (rank : 99) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HAU5, Q8N8U1, Q9H1J2, Q9NWL1, Q9P2D9, Q9Y4M9 | Gene names | UPF2, KIAA1408, RENT2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 2 (Nonsense mRNA reducing factor 2) (Up-frameshift suppressor 2 homolog) (hUpf2). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.040042 (rank : 137) | θ value | 8.99809 (rank : 101) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
MYO9B_HUMAN
|
||||||
| NC score | 0.036999 (rank : 138) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 641 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q13459, O75314, Q9NUJ2, Q9UHN0 | Gene names | MYO9B, MYR5 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9B (Myosin IXb) (Unconventional myosin-9b). | |||||
|
PHF22_HUMAN
|
||||||
| NC score | 0.035524 (rank : 139) | θ value | 4.03905 (rank : 75) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q96CB8, Q9HD71 | Gene names | INTS12, PHF22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Integrator complex subunit 12 (PHD finger protein 22). | |||||
|
CAR10_HUMAN
|
||||||
| NC score | 0.034473 (rank : 140) | θ value | 4.03905 (rank : 70) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 569 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BWT7, Q14CQ8, Q5TFG6, Q9UGR5, Q9UGR6, Q9Y3H0 | Gene names | CARD10, CARMA3 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (CARD-containing MAGUK protein 3) (Carma 3). | |||||
|
DDX21_HUMAN
|
||||||
| NC score | 0.033013 (rank : 141) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NR30, Q13436, Q5VX41, Q68D35 | Gene names | DDX21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
SASH1_HUMAN
|
||||||
| NC score | 0.031998 (rank : 142) | θ value | 3.0926 (rank : 65) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | O94885, Q8TAI0, Q9H7R7 | Gene names | SASH1, KIAA0790, PEPE1 | |||
|
Domain Architecture |

|
|||||
| Description | SAM and SH3 domain-containing protein 1 (Proline-glutamate repeat- containing protein). | |||||
|
ANK3_HUMAN
|
||||||
| NC score | 0.031185 (rank : 143) | θ value | 0.365318 (rank : 21) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 853 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q12955 | Gene names | ANK3 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin-3 (ANK-3) (Ankyrin-G). | |||||
|
RGS3_HUMAN
|
||||||
| NC score | 0.030839 (rank : 144) | θ value | 8.99809 (rank : 117) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1375 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P49796, Q5VXC1, Q5VZ05, Q6ZRM5, Q8IUQ1, Q8NC47, Q8NFN4, Q8NFN5, Q8TD59, Q8TD68, Q8WXA0 | Gene names | RGS3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (RGP3). | |||||
|
BRD2_HUMAN
|
||||||
| NC score | 0.030245 (rank : 145) | θ value | 4.03905 (rank : 68) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P25440, O00699, O00700, Q15310, Q5STC9, Q969U4 | Gene names | BRD2, KIAA9001, RING3 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 2 (Protein RING3) (O27.1.1). | |||||
|
PREB_MOUSE
|
||||||
| NC score | 0.029159 (rank : 146) | θ value | 2.36792 (rank : 54) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WUQ2, Q9QZ99 | Gene names | Preb, Sec12 | |||
|
Domain Architecture |

|
|||||
| Description | Prolactin regulatory element-binding protein (Mammalian guanine nucleotide exchange factor mSec12). | |||||
|
ABCF1_HUMAN
|
||||||
| NC score | 0.029080 (rank : 147) | θ value | 8.99809 (rank : 100) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 343 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8NE71, O14897, Q69YP6 | Gene names | ABCF1, ABC50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family F member 1 (ATP-binding cassette 50) (TNF-alpha-stimulated ABC protein). | |||||
|
MOR2A_MOUSE
|
||||||
| NC score | 0.028827 (rank : 148) | θ value | 1.81305 (rank : 46) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q69ZX6, Q5QNQ7, Q6P547 | Gene names | Morc2a, Kiaa0852, Zcwcc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MORC family CW-type zinc finger protein 2A (Zinc finger CW-type coiled-coil domain protein 1). | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.028270 (rank : 149) | θ value | 3.0926 (rank : 63) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
NCOR1_HUMAN
|
||||||
| NC score | 0.026979 (rank : 150) | θ value | 8.99809 (rank : 111) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O75376, Q9UPV5, Q9UQ18 | Gene names | NCOR1, KIAA1047 | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear receptor corepressor 1 (N-CoR1) (N-CoR). | |||||
|
CHD6_HUMAN
|
||||||
| NC score | 0.026642 (rank : 151) | θ value | 3.0926 (rank : 57) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TD26, Q5JYQ0, Q5TGZ9, Q5TH00, Q5TH01, Q8IZR2, Q8WTY0, Q9H4H6, Q9H6D4, Q9NTT7, Q9P2L1 | Gene names | CHD6, CHD5, KIAA1335, RIGB | |||
|
Domain Architecture |

|
|||||
| Description | Chromodomain-helicase-DNA-binding protein 6 (EC 3.6.1.-) (ATP- dependent helicase CHD6) (CHD-6) (Radiation-induced gene B protein). | |||||
|
CENPO_MOUSE
|
||||||
| NC score | 0.026477 (rank : 152) | θ value | 5.27518 (rank : 84) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8K015, Q8R587, Q9CYY6 | Gene names | Cenpo | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centromere protein O (CENP-O). | |||||
|
NUCL_MOUSE
|
||||||
| NC score | 0.026047 (rank : 153) | θ value | 5.27518 (rank : 90) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09405, Q548M9, Q61991, Q99K50 | Gene names | Ncl, Nuc | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolin (Protein C23). | |||||
|
FMNL_MOUSE
|
||||||
| NC score | 0.025946 (rank : 154) | θ value | 1.81305 (rank : 45) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9JL26, Q6KAN4, Q9Z2V7 | Gene names | Fmnl1, Frl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-like protein 1 (Formin-related protein). | |||||
|
NEBU_HUMAN
|
||||||
| NC score | 0.025540 (rank : 155) | θ value | 2.36792 (rank : 53) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 420 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P20929, Q15346 | Gene names | NEB | |||
|
Domain Architecture |

|
|||||
| Description | Nebulin. | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.025355 (rank : 156) | θ value | 2.36792 (rank : 51) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
PNRC1_HUMAN
|
||||||
| NC score | 0.025256 (rank : 157) | θ value | 8.99809 (rank : 116) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q12796, Q5T7J6 | Gene names | PNRC1, PROL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich nuclear receptor coactivator 1 (Proline-rich protein 2) (B4-2 protein). | |||||
|
NO40_MOUSE
|
||||||
| NC score | 0.023953 (rank : 158) | θ value | 8.99809 (rank : 112) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ESX4 | Gene names | Zcchc17, Ps1d | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleolar protein of 40 kDa (pNO40) (Zinc finger CCHC domain- containing protein 17) (Putative S1 RNA-binding domain protein) (PS1D protein). | |||||
|
PLCB1_HUMAN
|
||||||
| NC score | 0.022968 (rank : 159) | θ value | 8.99809 (rank : 114) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9NQ66, O60325, Q5TFF7, Q5TGC9, Q8IV93, Q9BQW2, Q9H4H2, Q9H8H5, Q9NQ65, Q9NQH9, Q9NTH4, Q9UJP6, Q9UM26 | Gene names | PLCB1, KIAA0581 | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.022136 (rank : 160) | θ value | 5.27518 (rank : 83) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
CAR10_MOUSE
|
||||||
| NC score | 0.021957 (rank : 161) | θ value | 8.99809 (rank : 103) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 407 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P58660 | Gene names | Card10, Bimp1 | |||
|
Domain Architecture |

|
|||||
| Description | Caspase recruitment domain-containing protein 10 (Bcl10-interacting MAGUK protein 1) (Bimp1). | |||||
|
BRE1A_HUMAN
|
||||||
| NC score | 0.020776 (rank : 162) | θ value | 8.99809 (rank : 102) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1071 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q5VTR2, Q69YL5, Q6P527, Q8N3J4, Q96JD3, Q9H9Y7, Q9HA51, Q9NUR4, Q9NWQ3, Q9NX83 | Gene names | RNF20, BRE1A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (hBRE1) (RING finger protein 20). | |||||
|
PLCB1_MOUSE
|
||||||
| NC score | 0.020720 (rank : 163) | θ value | 8.99809 (rank : 115) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 531 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9Z1B3, Q62075, Q8K5A5, Q8K5A6, Q9Z0E5, Q9Z2T5 | Gene names | Plcb1, Plcb | |||
|
Domain Architecture |

|
|||||
| Description | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 1 (EC 3.1.4.11) (Phosphoinositide phospholipase C) (Phospholipase C- beta-1) (PLC-beta-1) (PLC-I) (PLC-154). | |||||
|
CCD21_HUMAN
|
||||||
| NC score | 0.020603 (rank : 164) | θ value | 8.99809 (rank : 104) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1228 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q6P2H3, Q5VY68, Q5VY70, Q9H6Q1, Q9H828, Q9UF52 | Gene names | CCDC21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 21. | |||||
|
CCD27_MOUSE
|
||||||
| NC score | 0.019583 (rank : 165) | θ value | 6.88961 (rank : 95) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
ZO2_MOUSE
|
||||||
| NC score | 0.019395 (rank : 166) | θ value | 4.03905 (rank : 80) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9Z0U1, Q8K210 | Gene names | Tjp2, Zo2 | |||
|
Domain Architecture |

|
|||||
| Description | Tight junction protein ZO-2 (Zonula occludens 2 protein) (Zona occludens 2 protein) (Tight junction protein 2). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.019169 (rank : 167) | θ value | 5.27518 (rank : 82) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
PKHA6_HUMAN
|
||||||
| NC score | 0.018627 (rank : 168) | θ value | 6.88961 (rank : 98) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Y2H5 | Gene names | PLEKHA6, KIAA0969, PEPP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family A member 6 (Phosphoinositol 3-phosphate-binding protein 3) (PEPP-3). | |||||
|
MAGD2_HUMAN
|
||||||
| NC score | 0.018457 (rank : 169) | θ value | 3.0926 (rank : 60) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9UNF1, O76058, Q5BJF3, Q8NAL6, Q9H218, Q9P0U9, Q9UM52 | Gene names | MAGED2, BCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen D2 (MAGE-D2 antigen) (MAGE-D) (Breast cancer-associated gene 1 protein) (BCG-1) (11B6) (Hepatocellular carcinoma-associated protein JCL-1). | |||||
|
SPEG_MOUSE
|
||||||
| NC score | 0.018027 (rank : 170) | θ value | 4.03905 (rank : 79) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 1959 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q62407, Q3TPH8, Q6P5V1, Q80TF7, Q80ZN0, Q8BZF4, Q9EQJ5 | Gene names | Speg, Apeg1, Kiaa1297 | |||
|
Domain Architecture |

|
|||||
| Description | Striated muscle-specific serine/threonine protein kinase (EC 2.7.11.1) (Aortic preferentially expressed protein 1) (APEG-1). | |||||
|
CJ118_HUMAN
|
||||||
| NC score | 0.017239 (rank : 171) | θ value | 5.27518 (rank : 85) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
RYR3_HUMAN
|
||||||
| NC score | 0.017073 (rank : 172) | θ value | 4.03905 (rank : 78) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15413, O15175, Q15412 | Gene names | RYR3, HBRR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 3 (Brain-type ryanodine receptor) (RyR3) (RYR-3) (Brain ryanodine receptor-calcium release channel). | |||||
|
MUG1_MOUSE
|
||||||
| NC score | 0.016117 (rank : 173) | θ value | 3.0926 (rank : 62) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28665 | Gene names | Mug1, Mug-1 | |||
|
Domain Architecture |

|
|||||
| Description | Murinoglobulin-1 precursor (MuG1). | |||||
|
FGD6_MOUSE
|
||||||
| NC score | 0.014803 (rank : 174) | θ value | 5.27518 (rank : 87) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
MCF2L_MOUSE
|
||||||
| NC score | 0.013977 (rank : 175) | θ value | 5.27518 (rank : 89) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q64096 | Gene names | Mcf2l, Dbs | |||
|
Domain Architecture |

|
|||||
| Description | Guanine nucleotide exchange factor DBS (DBL's big sister) (MCF2- transforming sequence-like protein). | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.013693 (rank : 176) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
ROCK2_HUMAN
|
||||||
| NC score | 0.013407 (rank : 177) | θ value | 2.36792 (rank : 55) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2073 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O75116, Q9UQN5 | Gene names | ROCK2, KIAA0619 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2) (Rho kinase 2). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.013367 (rank : 178) | θ value | 8.99809 (rank : 118) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
HMGB2_HUMAN
|
||||||
| NC score | 0.012345 (rank : 179) | θ value | 8.99809 (rank : 107) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P26583 | Gene names | HMGB2, HMG2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
HXA13_MOUSE
|
||||||
| NC score | 0.012142 (rank : 180) | θ value | 3.0926 (rank : 59) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62424 | Gene names | Hoxa13, Hox-1.10 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A13 (Hox-1.10). | |||||
|
HMGB2_MOUSE
|
||||||
| NC score | 0.011303 (rank : 181) | θ value | 8.99809 (rank : 108) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P30681, Q3UXT1, Q9EQD5 | Gene names | Hmgb2, Hmg2 | |||
|
Domain Architecture |

|
|||||
| Description | High mobility group protein B2 (High mobility group protein 2) (HMG- 2). | |||||
|
CTRO_HUMAN
|
||||||
| NC score | 0.010357 (rank : 182) | θ value | 5.27518 (rank : 86) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 2092 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | O14578, Q6XUH8, Q86UQ9, Q9UPZ7 | Gene names | CIT, KIAA0949, STK21 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
HXA13_HUMAN
|
||||||
| NC score | 0.010311 (rank : 183) | θ value | 6.88961 (rank : 96) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P31271, O43371 | Gene names | HOXA13, HOX1J | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein Hox-A13 (Hox-1J). | |||||
|
CT075_MOUSE
|
||||||
| NC score | 0.007085 (rank : 184) | θ value | 8.99809 (rank : 105) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 358 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P59383 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Uncharacterized protein C20orf75 homolog precursor. | |||||
|
PCD19_HUMAN
|
||||||
| NC score | 0.004121 (rank : 185) | θ value | 6.88961 (rank : 97) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8TAB3, Q5JTG1, Q5JTG2, Q68DT7, Q9P2N3 | Gene names | PCDH19, KIAA1313 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-19 precursor. | |||||
|
PCD19_MOUSE
|
||||||
| NC score | 0.003929 (rank : 186) | θ value | 8.99809 (rank : 113) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q80TF3, Q8BL13 | Gene names | Pcdh19, Kiaa1313 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protocadherin-19 precursor. | |||||
|
ZN75A_HUMAN
|
||||||
| NC score | 0.001453 (rank : 187) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 720 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96N20, Q92669 | Gene names | ZNF75A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 75A. | |||||
|
CTCF_HUMAN
|
||||||
| NC score | -0.000050 (rank : 188) | θ value | 8.99809 (rank : 106) | |||
| Query Neighborhood Hits | 118 | Target Neighborhood Hits | 764 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P49711 | Gene names | CTCF | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional repressor CTCF (CCCTC-binding factor) (CTCFL paralog) (11-zinc finger protein). | |||||