Please be patient as the page loads
|
DCPS_MOUSE
|
||||||
| SwissProt Accessions | Q9DAR7, Q8C5I7 | Gene names | Dcps, Dcs1, Hint5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger mRNA decapping enzyme DcpS (EC 3.-.-.-) (DCS-1) (Hint- related 7meGMP-directed hydrolase) (Histidine triad protein member 5) (HINT-5). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
DCPS_MOUSE
|
||||||
| θ value | 5.36813e-170 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DAR7, Q8C5I7 | Gene names | Dcps, Dcs1, Hint5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger mRNA decapping enzyme DcpS (EC 3.-.-.-) (DCS-1) (Hint- related 7meGMP-directed hydrolase) (Histidine triad protein member 5) (HINT-5). | |||||
|
DCPS_HUMAN
|
||||||
| θ value | 1.41594e-162 (rank : 2) | NC score | 0.998146 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96C86, Q8NHL8, Q9Y2S5 | Gene names | DCPS, DCS1, HINT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger mRNA decapping enzyme DcpS (EC 3.-.-.-) (DCS-1) (Hint- related 7meGMP-directed hydrolase) (Histidine triad protein member 5) (HINT-5). | |||||
|
APTX_MOUSE
|
||||||
| θ value | 0.00298849 (rank : 3) | NC score | 0.218305 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TQC5, Q8BPA7, Q8C2B5, Q8K3D1, Q9CQ59 | Gene names | Aptx | |||
|
Domain Architecture |

|
|||||
| Description | Aprataxin homolog (Forkhead-associated domain histidine triad-like protein) (FHA-HIT). | |||||
|
APTX_HUMAN
|
||||||
| θ value | 0.00665767 (rank : 4) | NC score | 0.209625 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z2E3, Q5T781, Q7Z2F3, Q7Z336, Q7Z5R5, Q7Z6V7, Q7Z6V8, Q9NXM5 | Gene names | APTX | |||
|
Domain Architecture |

|
|||||
| Description | Aprataxin (Forkhead-associated domain histidine triad-like protein) (FHA-HIT). | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 5) | NC score | 0.067991 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
ROBO1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 6) | NC score | 0.020837 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6N7, Q7Z300, Q9BUS7 | Gene names | ROBO1, DUTT1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor (H-Robo-1) (Deleted in U twenty twenty). | |||||
|
EMSY_HUMAN
|
||||||
| θ value | 3.0926 (rank : 7) | NC score | 0.040440 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
EMSY_MOUSE
|
||||||
| θ value | 3.0926 (rank : 8) | NC score | 0.040390 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
RYR1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 9) | NC score | 0.021918 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
TRI59_HUMAN
|
||||||
| θ value | 5.27518 (rank : 10) | NC score | 0.010690 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IWR1 | Gene names | TRIM59, RNF104, TRIM57, TSBF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 59 (Tumor suppressor TSBF-1) (RING finger protein 104). | |||||
|
CP26B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 11) | NC score | 0.007310 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NR63, Q53TW1, Q9NP41 | Gene names | CYP26B1, CYP26A2, P450RAI2 | |||
|
Domain Architecture |
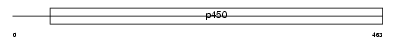
|
|||||
| Description | Cytochrome P450 26B1 (EC 1.14.-.-) (P450 26A2) (P450 retinoic acid- inactivating 2) (P450RAI-2) (Retinoic acid-metabolizing cytochrome). | |||||
|
ROBO1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 12) | NC score | 0.014269 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O89026 | Gene names | Robo1, Dutt1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor. | |||||
|
DCPS_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 5.36813e-170 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9DAR7, Q8C5I7 | Gene names | Dcps, Dcs1, Hint5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger mRNA decapping enzyme DcpS (EC 3.-.-.-) (DCS-1) (Hint- related 7meGMP-directed hydrolase) (Histidine triad protein member 5) (HINT-5). | |||||
|
DCPS_HUMAN
|
||||||
| NC score | 0.998146 (rank : 2) | θ value | 1.41594e-162 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q96C86, Q8NHL8, Q9Y2S5 | Gene names | DCPS, DCS1, HINT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Scavenger mRNA decapping enzyme DcpS (EC 3.-.-.-) (DCS-1) (Hint- related 7meGMP-directed hydrolase) (Histidine triad protein member 5) (HINT-5). | |||||
|
APTX_MOUSE
|
||||||
| NC score | 0.218305 (rank : 3) | θ value | 0.00298849 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7TQC5, Q8BPA7, Q8C2B5, Q8K3D1, Q9CQ59 | Gene names | Aptx | |||
|
Domain Architecture |

|
|||||
| Description | Aprataxin homolog (Forkhead-associated domain histidine triad-like protein) (FHA-HIT). | |||||
|
APTX_HUMAN
|
||||||
| NC score | 0.209625 (rank : 4) | θ value | 0.00665767 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z2E3, Q5T781, Q7Z2F3, Q7Z336, Q7Z5R5, Q7Z6V7, Q7Z6V8, Q9NXM5 | Gene names | APTX | |||
|
Domain Architecture |

|
|||||
| Description | Aprataxin (Forkhead-associated domain histidine triad-like protein) (FHA-HIT). | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.067991 (rank : 5) | θ value | 0.813845 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
EMSY_HUMAN
|
||||||
| NC score | 0.040440 (rank : 6) | θ value | 3.0926 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q7Z589, Q4G109, Q8NBU6, Q8TE50, Q9H8I9, Q9NRH0 | Gene names | EMSY, C11orf30 | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
EMSY_MOUSE
|
||||||
| NC score | 0.040390 (rank : 7) | θ value | 3.0926 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 294 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BMB0, Q5FWK5, Q80XU1, Q8VDW9 | Gene names | Emsy | |||
|
Domain Architecture |

|
|||||
| Description | Protein EMSY. | |||||
|
RYR1_HUMAN
|
||||||
| NC score | 0.021918 (rank : 8) | θ value | 5.27518 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21817, Q16314, Q16368, Q9NPK1, Q9P1U4 | Gene names | RYR1, RYDR | |||
|
Domain Architecture |

|
|||||
| Description | Ryanodine receptor 1 (Skeletal muscle-type ryanodine receptor) (RyR1) (RYR-1) (Skeletal muscle calcium release channel). | |||||
|
ROBO1_HUMAN
|
||||||
| NC score | 0.020837 (rank : 9) | θ value | 1.06291 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6N7, Q7Z300, Q9BUS7 | Gene names | ROBO1, DUTT1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor (H-Robo-1) (Deleted in U twenty twenty). | |||||
|
ROBO1_MOUSE
|
||||||
| NC score | 0.014269 (rank : 10) | θ value | 8.99809 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O89026 | Gene names | Robo1, Dutt1 | |||
|
Domain Architecture |

|
|||||
| Description | Roundabout homolog 1 precursor. | |||||
|
TRI59_HUMAN
|
||||||
| NC score | 0.010690 (rank : 11) | θ value | 5.27518 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IWR1 | Gene names | TRIM59, RNF104, TRIM57, TSBF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 59 (Tumor suppressor TSBF-1) (RING finger protein 104). | |||||
|
CP26B_HUMAN
|
||||||
| NC score | 0.007310 (rank : 12) | θ value | 8.99809 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NR63, Q53TW1, Q9NP41 | Gene names | CYP26B1, CYP26A2, P450RAI2 | |||
|
Domain Architecture |

|
|||||
| Description | Cytochrome P450 26B1 (EC 1.14.-.-) (P450 26A2) (P450 retinoic acid- inactivating 2) (P450RAI-2) (Retinoic acid-metabolizing cytochrome). | |||||