Please be patient as the page loads
|
CRLS1_HUMAN
|
||||||
| SwissProt Accessions | Q9UJA2, Q27RP0, Q69YQ5 | Gene names | CRLS1, C20orf155, CLS1 | |||
|
Domain Architecture |
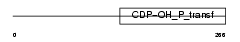
|
|||||
| Description | Cardiolipin synthetase (EC 2.7.8.-) (Cardiolipin synthase) (CLS) (Protein GCD10 homolog). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CRLS1_HUMAN
|
||||||
| θ value | 4.45295e-124 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UJA2, Q27RP0, Q69YQ5 | Gene names | CRLS1, C20orf155, CLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Cardiolipin synthetase (EC 2.7.8.-) (Cardiolipin synthase) (CLS) (Protein GCD10 homolog). | |||||
|
CRLS1_MOUSE
|
||||||
| θ value | 1.64275e-110 (rank : 2) | NC score | 0.997720 (rank : 2) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80ZM8, Q9D4U1 | Gene names | Crls1 | |||
|
Domain Architecture |
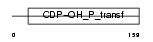
|
|||||
| Description | Cardiolipin synthetase (EC 2.7.8.-) (Cardiolipin synthase) (CLS). | |||||
|
CDIPT_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 3) | NC score | 0.179703 (rank : 3) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VDP6 | Gene names | Cdipt, Pis1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDP-diacylglycerol--inositol 3-phosphatidyltransferase (EC 2.7.8.11) (Phosphatidylinositol synthase) (PtdIns synthase) (PI synthase). | |||||
|
CDIPT_HUMAN
|
||||||
| θ value | 0.47712 (rank : 4) | NC score | 0.159322 (rank : 4) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14735, Q6FGU1, Q6ZN70 | Gene names | CDIPT, PIS, PIS1 | |||
|
Domain Architecture |
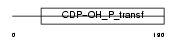
|
|||||
| Description | CDP-diacylglycerol--inositol 3-phosphatidyltransferase (EC 2.7.8.11) (Phosphatidylinositol synthase) (PtdIns synthase) (PI synthase). | |||||
|
PLCC_HUMAN
|
||||||
| θ value | 5.27518 (rank : 5) | NC score | 0.019982 (rank : 6) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRZ7, Q3ZCU2, Q6UWP6, Q6ZUC6, Q8N3Q7, Q9NRZ6 | Gene names | AGPAT3 | |||
|
Domain Architecture |
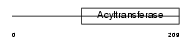
|
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase gamma (EC 2.3.1.51) (1- AGP acyltransferase 3) (1-AGPAT 3) (Lysophosphatidic acid acyltransferase-gamma) (LPAAT-gamma) (1-acylglycerol-3-phosphate O- acyltransferase 3). | |||||
|
ANKH_HUMAN
|
||||||
| θ value | 6.88961 (rank : 6) | NC score | 0.022587 (rank : 5) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HCJ1, Q9NQW2 | Gene names | ANKH, KIAA1581 | |||
|
Domain Architecture |
|
|||||
| Description | Progressive ankylosis protein homolog (ANK). | |||||
|
CELR2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 7) | NC score | 0.006154 (rank : 8) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
CELR2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 8) | NC score | 0.006109 (rank : 9) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||
|
PAX8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 9) | NC score | 0.008891 (rank : 7) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06710, Q09155, Q16337, Q16338, Q16339, Q96J49 | Gene names | PAX8 | |||
|
Domain Architecture |
|
|||||
| Description | Paired box protein Pax-8. | |||||
|
CRLS1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.45295e-124 (rank : 1) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9UJA2, Q27RP0, Q69YQ5 | Gene names | CRLS1, C20orf155, CLS1 | |||
|
Domain Architecture |

|
|||||
| Description | Cardiolipin synthetase (EC 2.7.8.-) (Cardiolipin synthase) (CLS) (Protein GCD10 homolog). | |||||
|
CRLS1_MOUSE
|
||||||
| NC score | 0.997720 (rank : 2) | θ value | 1.64275e-110 (rank : 2) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q80ZM8, Q9D4U1 | Gene names | Crls1 | |||
|
Domain Architecture |

|
|||||
| Description | Cardiolipin synthetase (EC 2.7.8.-) (Cardiolipin synthase) (CLS). | |||||
|
CDIPT_MOUSE
|
||||||
| NC score | 0.179703 (rank : 3) | θ value | 0.0961366 (rank : 3) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VDP6 | Gene names | Cdipt, Pis1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDP-diacylglycerol--inositol 3-phosphatidyltransferase (EC 2.7.8.11) (Phosphatidylinositol synthase) (PtdIns synthase) (PI synthase). | |||||
|
CDIPT_HUMAN
|
||||||
| NC score | 0.159322 (rank : 4) | θ value | 0.47712 (rank : 4) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14735, Q6FGU1, Q6ZN70 | Gene names | CDIPT, PIS, PIS1 | |||
|
Domain Architecture |

|
|||||
| Description | CDP-diacylglycerol--inositol 3-phosphatidyltransferase (EC 2.7.8.11) (Phosphatidylinositol synthase) (PtdIns synthase) (PI synthase). | |||||
|
ANKH_HUMAN
|
||||||
| NC score | 0.022587 (rank : 5) | θ value | 6.88961 (rank : 6) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HCJ1, Q9NQW2 | Gene names | ANKH, KIAA1581 | |||
|
Domain Architecture |

|
|||||
| Description | Progressive ankylosis protein homolog (ANK). | |||||
|
PLCC_HUMAN
|
||||||
| NC score | 0.019982 (rank : 6) | θ value | 5.27518 (rank : 5) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRZ7, Q3ZCU2, Q6UWP6, Q6ZUC6, Q8N3Q7, Q9NRZ6 | Gene names | AGPAT3 | |||
|
Domain Architecture |

|
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase gamma (EC 2.3.1.51) (1- AGP acyltransferase 3) (1-AGPAT 3) (Lysophosphatidic acid acyltransferase-gamma) (LPAAT-gamma) (1-acylglycerol-3-phosphate O- acyltransferase 3). | |||||
|
PAX8_HUMAN
|
||||||
| NC score | 0.008891 (rank : 7) | θ value | 8.99809 (rank : 9) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q06710, Q09155, Q16337, Q16338, Q16339, Q96J49 | Gene names | PAX8 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-8. | |||||
|
CELR2_HUMAN
|
||||||
| NC score | 0.006154 (rank : 8) | θ value | 6.88961 (rank : 7) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HCU4, Q92566 | Gene names | CELSR2, CDHF10, EGFL2, KIAA0279, MEGF3 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Epidermal growth factor-like 2) (Multiple epidermal growth factor-like domains 3) (Flamingo 1). | |||||
|
CELR2_MOUSE
|
||||||
| NC score | 0.006109 (rank : 9) | θ value | 8.99809 (rank : 8) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0M0, Q99K26, Q9Z2R4 | Gene names | Celsr2 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin EGF LAG seven-pass G-type receptor 2 precursor (Flamingo 1) (mFmi1). | |||||