Please be patient as the page loads
|
CRLD1_MOUSE
|
||||||
| SwissProt Accessions | Q8CGD2, Q8K018, Q99MM6 | Gene names | Crispld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich secretory protein LCCL domain-containing 1 precursor (CocoaCrisp). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CRLD1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999217 (rank : 2) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H336 | Gene names | CRISPLD1, CRISP10, LCRISP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich secretory protein LCCL domain-containing 1 precursor (LCCL domain-containing cysteine-rich secretory protein 1) (Cysteine- rich secretory protein 10) (CRISP-10) (Trypsin inhibitor Hl) (CocoaCrisp). | |||||
|
CRLD1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CGD2, Q8K018, Q99MM6 | Gene names | Crispld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich secretory protein LCCL domain-containing 1 precursor (CocoaCrisp). | |||||
|
CRLD2_HUMAN
|
||||||
| θ value | 4.1007e-178 (rank : 3) | NC score | 0.983435 (rank : 3) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H0B8, Q6P590, Q6UWH0, Q6UXC6, Q96IB1, Q96K61 | Gene names | CRISPLD2, CRISP11, LCRISP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich secretory protein LCCL domain-containing 2 precursor (LCCL domain-containing cysteine-rich secretory protein 2) (Cysteine- rich secretory protein 11) (CRISP-11). | |||||
|
GLIP1_HUMAN
|
||||||
| θ value | 5.40856e-29 (rank : 4) | NC score | 0.811304 (rank : 4) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48060, Q15409, Q969K2 | Gene names | GLIPR1, GLIPR, RTVP1 | |||
|
Domain Architecture |
|
|||||
| Description | Glioma pathogenesis-related protein 1 precursor (GliPR 1) (RTVP-1 protein). | |||||
|
GLIP1_MOUSE
|
||||||
| θ value | 3.62785e-25 (rank : 5) | NC score | 0.794331 (rank : 5) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CWG1 | Gene names | Glipr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma pathogenesis-related protein 1 precursor (GliPR 1). | |||||
|
CRIS3_HUMAN
|
||||||
| θ value | 4.9032e-22 (rank : 6) | NC score | 0.732246 (rank : 6) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54108, Q15512, Q5JW83, Q9H108 | Gene names | CRISP3 | |||
|
Domain Architecture |
|
|||||
| Description | Cysteine-rich secretory protein 3 precursor (CRISP-3) (SGP28 protein). | |||||
|
CRIS1_MOUSE
|
||||||
| θ value | 4.15078e-21 (rank : 7) | NC score | 0.728953 (rank : 7) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q03401 | Gene names | Crisp1, Aeg-1, Aeg1 | |||
|
Domain Architecture |
|
|||||
| Description | Cysteine-rich secretory protein 1 precursor (CRISP-1) (Acidic epididymal glycoprotein 1) (Sperm-coating glycoprotein 1) (SCP 1). | |||||
|
CRIS2_HUMAN
|
||||||
| θ value | 2.06002e-20 (rank : 8) | NC score | 0.719509 (rank : 9) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16562 | Gene names | CRISP2, TPX1 | |||
|
Domain Architecture |
|
|||||
| Description | Cysteine-rich secretory protein 2 precursor (CRISP-2) (Testis-specific protein TPX-1). | |||||
|
CRIS2_MOUSE
|
||||||
| θ value | 8.65492e-19 (rank : 9) | NC score | 0.714192 (rank : 10) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16563 | Gene names | Crisp2, Tpx-1, Tpx1 | |||
|
Domain Architecture |
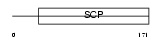
|
|||||
| Description | Cysteine-rich secretory protein 2 precursor (CRISP-2) (Testis-specific protein TPX-1). | |||||
|
CRIS1_HUMAN
|
||||||
| θ value | 7.32683e-18 (rank : 10) | NC score | 0.724924 (rank : 8) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54107, O00698, Q13248, Q14082, Q96SF6 | Gene names | CRISP1, AEGL1 | |||
|
Domain Architecture |
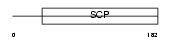
|
|||||
| Description | Cysteine-rich secretory protein 1 precursor (CRISP-1) (Acidic epididymal glycoprotein homolog) (AEG-like protein) (ARP). | |||||
|
CRIS3_MOUSE
|
||||||
| θ value | 1.68911e-14 (rank : 11) | NC score | 0.686539 (rank : 11) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q03402 | Gene names | Crisp3, Aeg-2, Aeg2 | |||
|
Domain Architecture |
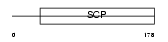
|
|||||
| Description | Cysteine-rich secretory protein 3 precursor (CRISP-3) (Acidic epididymal glycoprotein 2) (Sperm-coating glycoprotein 2) (SCP 2). | |||||
|
VITRN_MOUSE
|
||||||
| θ value | 1.06045e-08 (rank : 12) | NC score | 0.245993 (rank : 14) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHI5, Q3TZ47, Q8BQ41, Q8K047, Q9CYZ1 | Gene names | Vit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
VITRN_HUMAN
|
||||||
| θ value | 2.36244e-08 (rank : 13) | NC score | 0.244718 (rank : 15) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6UXI7, Q6P7T3, Q96DT1 | Gene names | VIT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
COCH_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 14) | NC score | 0.237780 (rank : 16) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43405 | Gene names | COCH, COCH5B2 | |||
|
Domain Architecture |

|
|||||
| Description | Cochlin precursor (COCH-5B2). | |||||
|
COCH_MOUSE
|
||||||
| θ value | 2.21117e-06 (rank : 15) | NC score | 0.229726 (rank : 17) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62507, Q3TAF5, Q9QWK6 | Gene names | Coch | |||
|
Domain Architecture |

|
|||||
| Description | Cochlin precursor (COCH-5B2). | |||||
|
DCBD2_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 16) | NC score | 0.148745 (rank : 20) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |
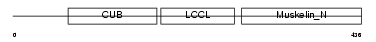
|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
DCBD2_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 17) | NC score | 0.143036 (rank : 21) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
DCBD1_MOUSE
|
||||||
| θ value | 0.00390308 (rank : 18) | NC score | 0.201685 (rank : 18) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DCBD1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 19) | NC score | 0.159606 (rank : 19) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
GAPR1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 20) | NC score | 0.370901 (rank : 12) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CYL5, Q3TJ42, Q8BTK4 | Gene names | Glipr2, Gapr1 | |||
|
Domain Architecture |
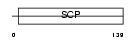
|
|||||
| Description | Golgi-associated plant pathogenesis-related protein 1 (Golgi- associated PR-1 protein) (GAPR-1) (Glioma pathogenesis-related protein 2) (GliPR 2). | |||||
|
LAMC3_MOUSE
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.011891 (rank : 26) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0B6, Q9WTW6 | Gene names | Lamc3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||
|
SELB_MOUSE
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.038478 (rank : 25) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JHW4 | Gene names | Eefsec, Selb | |||
|
Domain Architecture |

|
|||||
| Description | Selenocysteine-specific elongation factor (Elongation factor sec) (Eukaryotic elongation factor, selenocysteine-tRNA-specific) (mSelB). | |||||
|
IPPK_HUMAN
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.044076 (rank : 24) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H8X2, Q5T9F7, Q9H7V8 | Gene names | IPPK, C9orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-pentakisphosphate 2-kinase (EC 2.7.1.-) (Inositol-1,3,4,5,6- pentakisphosphate 2-kinase) (Ins(1,3,4,5,6)P5 2-kinase) (InsP5 2- kinase) (IPK1 homolog). | |||||
|
GAPR1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 24) | NC score | 0.331742 (rank : 13) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4G4, Q5VZR1, Q8N2S6, Q8WWC9, Q8WX36 | Gene names | GLIPR2, C9orf19, GAPR1 | |||
|
Domain Architecture |
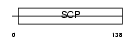
|
|||||
| Description | Golgi-associated plant pathogenesis-related protein 1 (Golgi- associated PR-1 protein) (GAPR-1) (Glioma pathogenesis-related protein 2) (GliPR 2). | |||||
|
MATN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.060537 (rank : 22) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21941 | Gene names | MATN1, CMP, CRTM | |||
|
Domain Architecture |
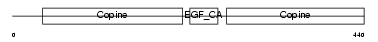
|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
MATN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.059440 (rank : 23) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51942 | Gene names | Matn1, Cmp, Crtm | |||
|
Domain Architecture |
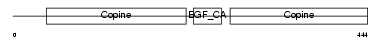
|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
CRLD1_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8CGD2, Q8K018, Q99MM6 | Gene names | Crispld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich secretory protein LCCL domain-containing 1 precursor (CocoaCrisp). | |||||
|
CRLD1_HUMAN
|
||||||
| NC score | 0.999217 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9H336 | Gene names | CRISPLD1, CRISP10, LCRISP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich secretory protein LCCL domain-containing 1 precursor (LCCL domain-containing cysteine-rich secretory protein 1) (Cysteine- rich secretory protein 10) (CRISP-10) (Trypsin inhibitor Hl) (CocoaCrisp). | |||||
|
CRLD2_HUMAN
|
||||||
| NC score | 0.983435 (rank : 3) | θ value | 4.1007e-178 (rank : 3) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H0B8, Q6P590, Q6UWH0, Q6UXC6, Q96IB1, Q96K61 | Gene names | CRISPLD2, CRISP11, LCRISP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cysteine-rich secretory protein LCCL domain-containing 2 precursor (LCCL domain-containing cysteine-rich secretory protein 2) (Cysteine- rich secretory protein 11) (CRISP-11). | |||||
|
GLIP1_HUMAN
|
||||||
| NC score | 0.811304 (rank : 4) | θ value | 5.40856e-29 (rank : 4) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P48060, Q15409, Q969K2 | Gene names | GLIPR1, GLIPR, RTVP1 | |||
|
Domain Architecture |

|
|||||
| Description | Glioma pathogenesis-related protein 1 precursor (GliPR 1) (RTVP-1 protein). | |||||
|
GLIP1_MOUSE
|
||||||
| NC score | 0.794331 (rank : 5) | θ value | 3.62785e-25 (rank : 5) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CWG1 | Gene names | Glipr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma pathogenesis-related protein 1 precursor (GliPR 1). | |||||
|
CRIS3_HUMAN
|
||||||
| NC score | 0.732246 (rank : 6) | θ value | 4.9032e-22 (rank : 6) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54108, Q15512, Q5JW83, Q9H108 | Gene names | CRISP3 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich secretory protein 3 precursor (CRISP-3) (SGP28 protein). | |||||
|
CRIS1_MOUSE
|
||||||
| NC score | 0.728953 (rank : 7) | θ value | 4.15078e-21 (rank : 7) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q03401 | Gene names | Crisp1, Aeg-1, Aeg1 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich secretory protein 1 precursor (CRISP-1) (Acidic epididymal glycoprotein 1) (Sperm-coating glycoprotein 1) (SCP 1). | |||||
|
CRIS1_HUMAN
|
||||||
| NC score | 0.724924 (rank : 8) | θ value | 7.32683e-18 (rank : 10) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P54107, O00698, Q13248, Q14082, Q96SF6 | Gene names | CRISP1, AEGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich secretory protein 1 precursor (CRISP-1) (Acidic epididymal glycoprotein homolog) (AEG-like protein) (ARP). | |||||
|
CRIS2_HUMAN
|
||||||
| NC score | 0.719509 (rank : 9) | θ value | 2.06002e-20 (rank : 8) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16562 | Gene names | CRISP2, TPX1 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich secretory protein 2 precursor (CRISP-2) (Testis-specific protein TPX-1). | |||||
|
CRIS2_MOUSE
|
||||||
| NC score | 0.714192 (rank : 10) | θ value | 8.65492e-19 (rank : 9) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P16563 | Gene names | Crisp2, Tpx-1, Tpx1 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich secretory protein 2 precursor (CRISP-2) (Testis-specific protein TPX-1). | |||||
|
CRIS3_MOUSE
|
||||||
| NC score | 0.686539 (rank : 11) | θ value | 1.68911e-14 (rank : 11) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q03402 | Gene names | Crisp3, Aeg-2, Aeg2 | |||
|
Domain Architecture |

|
|||||
| Description | Cysteine-rich secretory protein 3 precursor (CRISP-3) (Acidic epididymal glycoprotein 2) (Sperm-coating glycoprotein 2) (SCP 2). | |||||
|
GAPR1_MOUSE
|
||||||
| NC score | 0.370901 (rank : 12) | θ value | 0.279714 (rank : 20) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9CYL5, Q3TJ42, Q8BTK4 | Gene names | Glipr2, Gapr1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-associated plant pathogenesis-related protein 1 (Golgi- associated PR-1 protein) (GAPR-1) (Glioma pathogenesis-related protein 2) (GliPR 2). | |||||
|
GAPR1_HUMAN
|
||||||
| NC score | 0.331742 (rank : 13) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H4G4, Q5VZR1, Q8N2S6, Q8WWC9, Q8WX36 | Gene names | GLIPR2, C9orf19, GAPR1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgi-associated plant pathogenesis-related protein 1 (Golgi- associated PR-1 protein) (GAPR-1) (Glioma pathogenesis-related protein 2) (GliPR 2). | |||||
|
VITRN_MOUSE
|
||||||
| NC score | 0.245993 (rank : 14) | θ value | 1.06045e-08 (rank : 12) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHI5, Q3TZ47, Q8BQ41, Q8K047, Q9CYZ1 | Gene names | Vit | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
VITRN_HUMAN
|
||||||
| NC score | 0.244718 (rank : 15) | θ value | 2.36244e-08 (rank : 13) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q6UXI7, Q6P7T3, Q96DT1 | Gene names | VIT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vitrin precursor. | |||||
|
COCH_HUMAN
|
||||||
| NC score | 0.237780 (rank : 16) | θ value | 1.17247e-07 (rank : 14) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O43405 | Gene names | COCH, COCH5B2 | |||
|
Domain Architecture |

|
|||||
| Description | Cochlin precursor (COCH-5B2). | |||||
|
COCH_MOUSE
|
||||||
| NC score | 0.229726 (rank : 17) | θ value | 2.21117e-06 (rank : 15) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62507, Q3TAF5, Q9QWK6 | Gene names | Coch | |||
|
Domain Architecture |

|
|||||
| Description | Cochlin precursor (COCH-5B2). | |||||
|
DCBD1_MOUSE
|
||||||
| NC score | 0.201685 (rank : 18) | θ value | 0.00390308 (rank : 18) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9D4J3, Q8R327, Q9D696 | Gene names | Dcbld1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DCBD1_HUMAN
|
||||||
| NC score | 0.159606 (rank : 19) | θ value | 0.00869519 (rank : 19) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8N8Z6, Q5H992, Q8IYK5, Q8N7L9, Q96NH2 | Gene names | DCBLD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 1 precursor. | |||||
|
DCBD2_MOUSE
|
||||||
| NC score | 0.148745 (rank : 20) | θ value | 0.00020696 (rank : 16) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q91ZV3, Q8BKI4 | Gene names | Dcbld2, Esdn | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein). | |||||
|
DCBD2_HUMAN
|
||||||
| NC score | 0.143036 (rank : 21) | θ value | 0.00102713 (rank : 17) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q96PD2, Q8N6M4, Q8TDX2 | Gene names | DCBLD2, CLCP1, ESDN | |||
|
Domain Architecture |

|
|||||
| Description | Discoidin, CUB and LCCL domain-containing protein 2 precursor (Endothelial and smooth muscle cell-derived neuropilin-like protein) (CUB, LCCL and coagulation factor V/VIII-homology domains protein 1). | |||||
|
MATN1_HUMAN
|
||||||
| NC score | 0.060537 (rank : 22) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21941 | Gene names | MATN1, CMP, CRTM | |||
|
Domain Architecture |

|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
MATN1_MOUSE
|
||||||
| NC score | 0.059440 (rank : 23) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51942 | Gene names | Matn1, Cmp, Crtm | |||
|
Domain Architecture |

|
|||||
| Description | Cartilage matrix protein precursor (Matrilin-1). | |||||
|
IPPK_HUMAN
|
||||||
| NC score | 0.044076 (rank : 24) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9H8X2, Q5T9F7, Q9H7V8 | Gene names | IPPK, C9orf12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Inositol-pentakisphosphate 2-kinase (EC 2.7.1.-) (Inositol-1,3,4,5,6- pentakisphosphate 2-kinase) (Ins(1,3,4,5,6)P5 2-kinase) (InsP5 2- kinase) (IPK1 homolog). | |||||
|
SELB_MOUSE
|
||||||
| NC score | 0.038478 (rank : 25) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9JHW4 | Gene names | Eefsec, Selb | |||
|
Domain Architecture |

|
|||||
| Description | Selenocysteine-specific elongation factor (Elongation factor sec) (Eukaryotic elongation factor, selenocysteine-tRNA-specific) (mSelB). | |||||
|
LAMC3_MOUSE
|
||||||
| NC score | 0.011891 (rank : 26) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 624 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9R0B6, Q9WTW6 | Gene names | Lamc3 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin gamma-3 chain precursor (Laminin 12 gamma 3 subunit). | |||||