Please be patient as the page loads
|
COQ3_MOUSE
|
||||||
| SwissProt Accessions | Q8BMS4, Q3TCX5, Q6P918, Q8BM28 | Gene names | Coq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hexaprenyldihydroxybenzoate methyltransferase, mitochondrial precursor (EC 2.1.1.114) (Dihydroxyhexaprenylbenzoate methyltransferase) (3,4- dihydroxy-5-hexaprenylbenzoate methyltransferase) (DHHB methyltransferase) (DHHB-MT) (DHHB-MTase). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
COQ3_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BMS4, Q3TCX5, Q6P918, Q8BM28 | Gene names | Coq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hexaprenyldihydroxybenzoate methyltransferase, mitochondrial precursor (EC 2.1.1.114) (Dihydroxyhexaprenylbenzoate methyltransferase) (3,4- dihydroxy-5-hexaprenylbenzoate methyltransferase) (DHHB methyltransferase) (DHHB-MT) (DHHB-MTase). | |||||
|
COQ3_HUMAN
|
||||||
| θ value | 5.96274e-153 (rank : 2) | NC score | 0.987828 (rank : 2) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NZJ6, Q6P4F0, Q8IXG6, Q96BG1, Q9H0N1 | Gene names | COQ3 | |||
|
Domain Architecture |
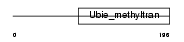
|
|||||
| Description | Hexaprenyldihydroxybenzoate methyltransferase, mitochondrial precursor (EC 2.1.1.114) (Dihydroxyhexaprenylbenzoate methyltransferase) (3,4- dihydroxy-5-hexaprenylbenzoate methyltransferase) (DHHB methyltransferase) (DHHB-MT) (DHHB-MTase). | |||||
|
AS3MT_MOUSE
|
||||||
| θ value | 0.00134147 (rank : 3) | NC score | 0.219348 (rank : 3) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91WU5, Q9QZT1 | Gene names | As3mt, Cyt19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arsenite methyltransferase (EC 2.1.1.137) (S-adenosyl-L- methionine:arsenic(III) methyltransferase) (Methylarsonite methyltransferase). | |||||
|
METL2_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 4) | NC score | 0.148907 (rank : 5) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96IZ6, Q9H9G9, Q9NUI8, Q9P0B5 | Gene names | METTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Methyltransferase-like protein 2 (EC 2.1.1.-). | |||||
|
AS3MT_HUMAN
|
||||||
| θ value | 0.0431538 (rank : 5) | NC score | 0.197507 (rank : 4) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HBK9 | Gene names | AS3MT, CYT19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arsenite methyltransferase (EC 2.1.1.137) (S-adenosyl-L- methionine:arsenic(III) methyltransferase) (Methylarsonite methyltransferase). | |||||
|
METL2_MOUSE
|
||||||
| θ value | 0.21417 (rank : 6) | NC score | 0.116146 (rank : 7) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BMK1, Q8BXC2 | Gene names | Mettl2, D11Ertd768e | |||
|
Domain Architecture |

|
|||||
| Description | Methyltransferase-like protein 2 (EC 2.1.1.-). | |||||
|
PIMT_HUMAN
|
||||||
| θ value | 0.365318 (rank : 7) | NC score | 0.112198 (rank : 8) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22061, Q14661, Q16556, Q93061, Q96II9, Q99625 | Gene names | PCMT1 | |||
|
Domain Architecture |
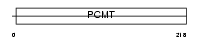
|
|||||
| Description | Protein-L-isoaspartate(D-aspartate) O-methyltransferase (EC 2.1.1.77) (Protein-beta-aspartate methyltransferase) (PIMT) (Protein L- isoaspartyl/D-aspartyl methyltransferase) (L-isoaspartyl protein carboxyl methyltransferase). | |||||
|
PIMT_MOUSE
|
||||||
| θ value | 0.365318 (rank : 8) | NC score | 0.129240 (rank : 6) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23506, Q810R4 | Gene names | Pcmt1 | |||
|
Domain Architecture |
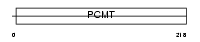
|
|||||
| Description | Protein-L-isoaspartate(D-aspartate) O-methyltransferase (EC 2.1.1.77) (Protein-beta-aspartate methyltransferase) (PIMT) (Protein L- isoaspartyl/D-aspartyl methyltransferase) (L-isoaspartyl protein carboxyl methyltransferase). | |||||
|
GNMT_HUMAN
|
||||||
| θ value | 0.62314 (rank : 9) | NC score | 0.097414 (rank : 9) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14749, Q9NNZ1, Q9NS24 | Gene names | GNMT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glycine N-methyltransferase (EC 2.1.1.20). | |||||
|
CI032_MOUSE
|
||||||
| θ value | 1.06291 (rank : 10) | NC score | 0.053702 (rank : 12) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R2U4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0351 protein C9orf32 homolog. | |||||
|
DPP4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 11) | NC score | 0.017487 (rank : 15) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27487 | Gene names | DPP4, ADCP2, CD26 | |||
|
Domain Architecture |

|
|||||
| Description | Dipeptidyl peptidase 4 (EC 3.4.14.5) (Dipeptidyl peptidase IV) (DPP IV) (T-cell activation antigen CD26) (TP103) (Adenosine deaminase complexing protein 2) (ADABP) [Contains: Dipeptidyl peptidase 4 membrane form (Dipeptidyl peptidase IV membrane form); Dipeptidyl peptidase 4 soluble form (Dipeptidyl peptidase IV soluble form)]. | |||||
|
COQ5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 12) | NC score | 0.031194 (rank : 14) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5HYK3, Q32Q28, Q53HH0, Q96LV1, Q9BSP8 | Gene names | COQ5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquinone biosynthesis methyltransferase COQ5, mitochondrial precursor (EC 2.1.1.-). | |||||
|
METL5_HUMAN
|
||||||
| θ value | 4.03905 (rank : 13) | NC score | 0.032960 (rank : 13) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRN9, Q9NVX1 | Gene names | METTL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyltransferase-like protein 5 (EC 2.1.1.-). | |||||
|
WBS27_HUMAN
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.071378 (rank : 10) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N6F8 | Gene names | WBSCR27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 27 protein. | |||||
|
MET7A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 15) | NC score | 0.066657 (rank : 11) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H8H3, Q9H7R3, Q9UHZ7, Q9Y422 | Gene names | METTL7A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyltransferase-like protein 7A precursor (EC 2.1.1.-) (Protein AAM- B). | |||||
|
COQ3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8BMS4, Q3TCX5, Q6P918, Q8BM28 | Gene names | Coq3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hexaprenyldihydroxybenzoate methyltransferase, mitochondrial precursor (EC 2.1.1.114) (Dihydroxyhexaprenylbenzoate methyltransferase) (3,4- dihydroxy-5-hexaprenylbenzoate methyltransferase) (DHHB methyltransferase) (DHHB-MT) (DHHB-MTase). | |||||
|
COQ3_HUMAN
|
||||||
| NC score | 0.987828 (rank : 2) | θ value | 5.96274e-153 (rank : 2) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9NZJ6, Q6P4F0, Q8IXG6, Q96BG1, Q9H0N1 | Gene names | COQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Hexaprenyldihydroxybenzoate methyltransferase, mitochondrial precursor (EC 2.1.1.114) (Dihydroxyhexaprenylbenzoate methyltransferase) (3,4- dihydroxy-5-hexaprenylbenzoate methyltransferase) (DHHB methyltransferase) (DHHB-MT) (DHHB-MTase). | |||||
|
AS3MT_MOUSE
|
||||||
| NC score | 0.219348 (rank : 3) | θ value | 0.00134147 (rank : 3) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91WU5, Q9QZT1 | Gene names | As3mt, Cyt19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arsenite methyltransferase (EC 2.1.1.137) (S-adenosyl-L- methionine:arsenic(III) methyltransferase) (Methylarsonite methyltransferase). | |||||
|
AS3MT_HUMAN
|
||||||
| NC score | 0.197507 (rank : 4) | θ value | 0.0431538 (rank : 5) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9HBK9 | Gene names | AS3MT, CYT19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arsenite methyltransferase (EC 2.1.1.137) (S-adenosyl-L- methionine:arsenic(III) methyltransferase) (Methylarsonite methyltransferase). | |||||
|
METL2_HUMAN
|
||||||
| NC score | 0.148907 (rank : 5) | θ value | 0.0252991 (rank : 4) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96IZ6, Q9H9G9, Q9NUI8, Q9P0B5 | Gene names | METTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Methyltransferase-like protein 2 (EC 2.1.1.-). | |||||
|
PIMT_MOUSE
|
||||||
| NC score | 0.129240 (rank : 6) | θ value | 0.365318 (rank : 8) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P23506, Q810R4 | Gene names | Pcmt1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-L-isoaspartate(D-aspartate) O-methyltransferase (EC 2.1.1.77) (Protein-beta-aspartate methyltransferase) (PIMT) (Protein L- isoaspartyl/D-aspartyl methyltransferase) (L-isoaspartyl protein carboxyl methyltransferase). | |||||
|
METL2_MOUSE
|
||||||
| NC score | 0.116146 (rank : 7) | θ value | 0.21417 (rank : 6) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BMK1, Q8BXC2 | Gene names | Mettl2, D11Ertd768e | |||
|
Domain Architecture |

|
|||||
| Description | Methyltransferase-like protein 2 (EC 2.1.1.-). | |||||
|
PIMT_HUMAN
|
||||||
| NC score | 0.112198 (rank : 8) | θ value | 0.365318 (rank : 7) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22061, Q14661, Q16556, Q93061, Q96II9, Q99625 | Gene names | PCMT1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein-L-isoaspartate(D-aspartate) O-methyltransferase (EC 2.1.1.77) (Protein-beta-aspartate methyltransferase) (PIMT) (Protein L- isoaspartyl/D-aspartyl methyltransferase) (L-isoaspartyl protein carboxyl methyltransferase). | |||||
|
GNMT_HUMAN
|
||||||
| NC score | 0.097414 (rank : 9) | θ value | 0.62314 (rank : 9) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14749, Q9NNZ1, Q9NS24 | Gene names | GNMT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glycine N-methyltransferase (EC 2.1.1.20). | |||||
|
WBS27_HUMAN
|
||||||
| NC score | 0.071378 (rank : 10) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N6F8 | Gene names | WBSCR27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Williams-Beuren syndrome chromosome region 27 protein. | |||||
|
MET7A_HUMAN
|
||||||
| NC score | 0.066657 (rank : 11) | θ value | 8.99809 (rank : 15) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H8H3, Q9H7R3, Q9UHZ7, Q9Y422 | Gene names | METTL7A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyltransferase-like protein 7A precursor (EC 2.1.1.-) (Protein AAM- B). | |||||
|
CI032_MOUSE
|
||||||
| NC score | 0.053702 (rank : 12) | θ value | 1.06291 (rank : 10) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8R2U4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UPF0351 protein C9orf32 homolog. | |||||
|
METL5_HUMAN
|
||||||
| NC score | 0.032960 (rank : 13) | θ value | 4.03905 (rank : 13) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NRN9, Q9NVX1 | Gene names | METTL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyltransferase-like protein 5 (EC 2.1.1.-). | |||||
|
COQ5_HUMAN
|
||||||
| NC score | 0.031194 (rank : 14) | θ value | 4.03905 (rank : 12) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5HYK3, Q32Q28, Q53HH0, Q96LV1, Q9BSP8 | Gene names | COQ5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquinone biosynthesis methyltransferase COQ5, mitochondrial precursor (EC 2.1.1.-). | |||||
|
DPP4_HUMAN
|
||||||
| NC score | 0.017487 (rank : 15) | θ value | 3.0926 (rank : 11) | |||
| Query Neighborhood Hits | 15 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27487 | Gene names | DPP4, ADCP2, CD26 | |||
|
Domain Architecture |

|
|||||
| Description | Dipeptidyl peptidase 4 (EC 3.4.14.5) (Dipeptidyl peptidase IV) (DPP IV) (T-cell activation antigen CD26) (TP103) (Adenosine deaminase complexing protein 2) (ADABP) [Contains: Dipeptidyl peptidase 4 membrane form (Dipeptidyl peptidase IV membrane form); Dipeptidyl peptidase 4 soluble form (Dipeptidyl peptidase IV soluble form)]. | |||||