Please be patient as the page loads
|
CM003_MOUSE
|
||||||
| SwissProt Accessions | Q8C263 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf3 homolog. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CM003_MOUSE
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8C263 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf3 homolog. | |||||
|
CM003_HUMAN
|
||||||
| θ value | 8.35901e-139 (rank : 2) | NC score | 0.967905 (rank : 2) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IX90, Q5VZV5, Q86WR2, Q8NBG1, Q96D22 | Gene names | C13orf3, RAMA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf3. | |||||
|
MAGC1_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 3) | NC score | 0.036243 (rank : 6) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
NEMO_HUMAN
|
||||||
| θ value | 0.21417 (rank : 4) | NC score | 0.028877 (rank : 8) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6K9 | Gene names | IKBKG, FIP3, NEMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (FIP-3). | |||||
|
ERCC5_HUMAN
|
||||||
| θ value | 0.813845 (rank : 5) | NC score | 0.047570 (rank : 4) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 6) | NC score | 0.056140 (rank : 3) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
EVI2B_MOUSE
|
||||||
| θ value | 1.81305 (rank : 7) | NC score | 0.045389 (rank : 5) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VD58, Q3U1A3 | Gene names | Evi2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EVI2B protein precursor (Ecotropic viral integration site 2B protein). | |||||
|
NACAM_MOUSE
|
||||||
| θ value | 1.81305 (rank : 8) | NC score | 0.023544 (rank : 13) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MILK1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 9) | NC score | 0.020833 (rank : 15) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
PCD15_HUMAN
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.009954 (rank : 20) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
BRD7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 11) | NC score | 0.030114 (rank : 7) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
JHD2A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 12) | NC score | 0.017278 (rank : 17) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 13) | NC score | 0.021034 (rank : 14) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
BRD7_MOUSE
|
||||||
| θ value | 6.88961 (rank : 14) | NC score | 0.025568 (rank : 10) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88665, Q3UQ56, Q3UU06, Q9CT78 | Gene names | Brd7, Bp75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein). | |||||
|
MIS_HUMAN
|
||||||
| θ value | 6.88961 (rank : 15) | NC score | 0.018425 (rank : 16) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P03971, O75246 | Gene names | AMH, MIF | |||
|
Domain Architecture |

|
|||||
| Description | Muellerian-inhibiting factor precursor (MIS) (Anti-Muellerian hormone) (AMH) (Muellerian-inhibiting substance). | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.007562 (rank : 21) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
PO121_MOUSE
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.027731 (rank : 9) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
RP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 18) | NC score | 0.015977 (rank : 19) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56716 | Gene names | Rp1, Orp1, Rp1h | |||
|
Domain Architecture |
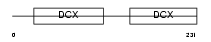
|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein homolog). | |||||
|
VP26A_HUMAN
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.024520 (rank : 12) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75436, Q8TBH4, Q9H982 | Gene names | VPS26A, VPS26 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 26A (Vesicle protein sorting 26A) (hVPS26). | |||||
|
VP26A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.024539 (rank : 11) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P40336, Q3TGY3, Q3THM5, Q3TW99, Q3UD54, Q8C1E9 | Gene names | Vps26a, Vps26 | |||
|
Domain Architecture |

|
|||||
| Description |
Vacuolar protein sorting-associated protein 26A (Vesicle protein
sorting 26A) (mVPS26) (H |
|||||
|
CP135_MOUSE
|
||||||
| θ value | 8.99809 (rank : 21) | NC score | 0.005161 (rank : 22) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
MUC2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.016515 (rank : 18) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
WNK2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.002344 (rank : 23) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||
|
CM003_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8C263 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf3 homolog. | |||||
|
CM003_HUMAN
|
||||||
| NC score | 0.967905 (rank : 2) | θ value | 8.35901e-139 (rank : 2) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8IX90, Q5VZV5, Q86WR2, Q8NBG1, Q96D22 | Gene names | C13orf3, RAMA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C13orf3. | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.056140 (rank : 3) | θ value | 0.813845 (rank : 6) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
ERCC5_HUMAN
|
||||||
| NC score | 0.047570 (rank : 4) | θ value | 0.813845 (rank : 5) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P28715, Q7Z2V3, Q8IZL6, Q8N1B7, Q9HD59 | Gene names | ERCC5, XPG, XPGC | |||
|
Domain Architecture |

|
|||||
| Description | DNA-repair protein complementing XP-G cells (Xeroderma pigmentosum group G-complementing protein) (DNA excision repair protein ERCC-5). | |||||
|
EVI2B_MOUSE
|
||||||
| NC score | 0.045389 (rank : 5) | θ value | 1.81305 (rank : 7) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VD58, Q3U1A3 | Gene names | Evi2b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EVI2B protein precursor (Ecotropic viral integration site 2B protein). | |||||
|
MAGC1_HUMAN
|
||||||
| NC score | 0.036243 (rank : 6) | θ value | 0.0961366 (rank : 3) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O60732, O75451, Q8TCV4 | Gene names | MAGEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Melanoma-associated antigen C1 (MAGE-C1 antigen) (Cancer-testis antigen CT7). | |||||
|
BRD7_HUMAN
|
||||||
| NC score | 0.030114 (rank : 7) | θ value | 3.0926 (rank : 11) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 94 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NPI1, Q4VC09, Q8N2L9, Q96KA4, Q9BV48, Q9UH59 | Gene names | BRD7, BP75, CELTIX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein) (Protein CELTIX-1). | |||||
|
NEMO_HUMAN
|
||||||
| NC score | 0.028877 (rank : 8) | θ value | 0.21417 (rank : 4) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 820 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6K9 | Gene names | IKBKG, FIP3, NEMO | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NF-kappa-B essential modulator (NEMO) (NF-kappa-B essential modifier) (Inhibitor of nuclear factor kappa-B kinase subunit gamma) (IkB kinase subunit gamma) (I-kappa-B kinase gamma) (IKK-gamma) (IKKG) (IkB kinase-associated protein 1) (IKKAP1) (FIP-3). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.027731 (rank : 9) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
BRD7_MOUSE
|
||||||
| NC score | 0.025568 (rank : 10) | θ value | 6.88961 (rank : 14) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O88665, Q3UQ56, Q3UU06, Q9CT78 | Gene names | Brd7, Bp75 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 7 (75 kDa bromodomain protein). | |||||
|
VP26A_MOUSE
|
||||||
| NC score | 0.024539 (rank : 11) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P40336, Q3TGY3, Q3THM5, Q3TW99, Q3UD54, Q8C1E9 | Gene names | Vps26a, Vps26 | |||
|
Domain Architecture |

|
|||||
| Description |
Vacuolar protein sorting-associated protein 26A (Vesicle protein
sorting 26A) (mVPS26) (H |
|||||
|
VP26A_HUMAN
|
||||||
| NC score | 0.024520 (rank : 12) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O75436, Q8TBH4, Q9H982 | Gene names | VPS26A, VPS26 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 26A (Vesicle protein sorting 26A) (hVPS26). | |||||
|
NACAM_MOUSE
|
||||||
| NC score | 0.023544 (rank : 13) | θ value | 1.81305 (rank : 8) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1866 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P70670 | Gene names | Naca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (Alpha-NAC, muscle-specific form). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.021034 (rank : 14) | θ value | 4.03905 (rank : 13) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MILK1_HUMAN
|
||||||
| NC score | 0.020833 (rank : 15) | θ value | 2.36792 (rank : 9) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N3F8, Q5TI16, Q7RTP5, Q8N3N8, Q9BVL9, Q9BY92, Q9UH43, Q9UH44, Q9UH45 | Gene names | MIRAB13, KIAA1668 | |||
|
Domain Architecture |

|
|||||
| Description | Molecule interacting with Rab13 (MIRab13) (MICAL-like protein 1). | |||||
|
MIS_HUMAN
|
||||||
| NC score | 0.018425 (rank : 16) | θ value | 6.88961 (rank : 15) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P03971, O75246 | Gene names | AMH, MIF | |||
|
Domain Architecture |

|
|||||
| Description | Muellerian-inhibiting factor precursor (MIS) (Anti-Muellerian hormone) (AMH) (Muellerian-inhibiting substance). | |||||
|
JHD2A_HUMAN
|
||||||
| NC score | 0.017278 (rank : 17) | θ value | 4.03905 (rank : 12) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Y4C1, Q53S72, Q68D47, Q68UT9, Q6N050, Q8IY08 | Gene names | JMJD1A, JHDM2A, JMJD1, KIAA0742 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 2A (EC 1.14.11.-) (Jumonji domain-containing protein 1A). | |||||
|
MUC2_HUMAN
|
||||||
| NC score | 0.016515 (rank : 18) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q02817, Q14878 | Gene names | MUC2, SMUC | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-2 precursor (Intestinal mucin-2). | |||||
|
RP1_MOUSE
|
||||||
| NC score | 0.015977 (rank : 19) | θ value | 6.88961 (rank : 18) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 104 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56716 | Gene names | Rp1, Orp1, Rp1h | |||
|
Domain Architecture |

|
|||||
| Description | Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein homolog). | |||||
|
PCD15_HUMAN
|
||||||
| NC score | 0.009954 (rank : 20) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96QU1, Q96QT8 | Gene names | PCDH15 | |||
|
Domain Architecture |

|
|||||
| Description | Protocadherin-15 precursor. | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.007562 (rank : 21) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.005161 (rank : 22) | θ value | 8.99809 (rank : 21) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
WNK2_HUMAN
|
||||||
| NC score | 0.002344 (rank : 23) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 23 | Target Neighborhood Hits | 1407 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y3S1, Q8IY36, Q9C0A3, Q9H3P4 | Gene names | WNK2, KIAA1760, PRKWNK2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK2 (EC 2.7.11.1) (Protein kinase with no lysine 2) (Protein kinase, lysine-deficient 2). | |||||