Please be patient as the page loads
|
CDS2_MOUSE
|
||||||
| SwissProt Accessions | Q99L43, Q3TMD1, Q6NSU1 | Gene names | Cds2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidate cytidylyltransferase 2 (EC 2.7.7.41) (CDP-diglyceride synthetase 2) (CDP-diglyceride pyrophosphorylase 2) (CDP- diacylglycerol synthase 2) (CDS 2) (CTP:phosphatidate cytidylyltransferase 2) (CDP-DAG synthase 2) (CDP-DG synthetase 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CDS1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.993455 (rank : 4) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92903, O00163 | Gene names | CDS1, CDS | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidate cytidylyltransferase 1 (EC 2.7.7.41) (CDP-diglyceride synthetase 1) (CDP-diglyceride pyrophosphorylase 1) (CDP- diacylglycerol synthase 1) (CDS 1) (CTP:phosphatidate cytidylyltransferase 1) (CDP-DAG synthase 1) (CDP-DG synthetase 1). | |||||
|
CDS1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994179 (rank : 3) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98191, Q8CGZ1 | Gene names | Cds1, Cds | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidate cytidylyltransferase 1 (EC 2.7.7.41) (CDP-diglyceride synthetase 1) (CDP-diglyceride pyrophosphorylase 1) (CDP- diacylglycerol synthase 1) (CDS 1) (CTP:phosphatidate cytidylyltransferase 1) (CDP-DAG synthase 1) (CDP-DG synthetase 1). | |||||
|
CDS2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.998698 (rank : 2) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95674, Q5TDY2, Q5TDY3, Q5TDY4, Q5TDY5, Q9BYK5, Q9NTT2 | Gene names | CDS2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidate cytidylyltransferase 2 (EC 2.7.7.41) (CDP-diglyceride synthetase 2) (CDP-diglyceride pyrophosphorylase 2) (CDP- diacylglycerol synthase 2) (CDS 2) (CTP:phosphatidate cytidylyltransferase 2) (CDP-DAG synthase 2) (CDP-DG synthetase 2). | |||||
|
CDS2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99L43, Q3TMD1, Q6NSU1 | Gene names | Cds2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidate cytidylyltransferase 2 (EC 2.7.7.41) (CDP-diglyceride synthetase 2) (CDP-diglyceride pyrophosphorylase 2) (CDP- diacylglycerol synthase 2) (CDS 2) (CTP:phosphatidate cytidylyltransferase 2) (CDP-DAG synthase 2) (CDP-DG synthetase 2). | |||||
|
G109A_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 5) | NC score | 0.039297 (rank : 9) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TDS4 | Gene names | GPR109A, HM74A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nicotinic acid receptor 1 (G-protein coupled receptor 109A) (G-protein coupled receptor HM74A). | |||||
|
G109B_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 6) | NC score | 0.042703 (rank : 7) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 579 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49019, Q8NGE4 | Gene names | GPR109B | |||
|
Domain Architecture |
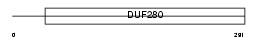
|
|||||
| Description | Nicotinic acid receptor 2 (G-protein coupled receptor 109B) (G-protein coupled receptor HM74) (G-protein coupled receptor HM74B). | |||||
|
ELOV4_HUMAN
|
||||||
| θ value | 0.163984 (rank : 7) | NC score | 0.064467 (rank : 5) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZR5, Q5TCS2, Q86YJ1, Q9H139 | Gene names | ELOVL4 | |||
|
Domain Architecture |
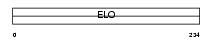
|
|||||
| Description | Elongation of very long chain fatty acids protein 4. | |||||
|
PARG_MOUSE
|
||||||
| θ value | 0.21417 (rank : 8) | NC score | 0.057376 (rank : 6) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88622, Q80YQ6, Q8CB72 | Gene names | Parg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
GP109_MOUSE
|
||||||
| θ value | 2.36792 (rank : 9) | NC score | 0.020814 (rank : 13) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EP66 | Gene names | Gpr109, Gpr109a, Gpr109b, Pumag | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nicotinic acid receptor (G-protein coupled receptor 109) (G-protein coupled receptor HM74) (Protein PUMA-G). | |||||
|
NPT1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 10) | NC score | 0.021751 (rank : 12) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14916, Q13783 | Gene names | SLC17A1, NPT1 | |||
|
Domain Architecture |
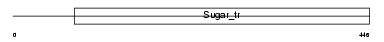
|
|||||
| Description | Sodium-dependent phosphate transport protein 1 (Sodium/phosphate cotransporter 1) (Na(+)/PI cotransporter 1) (Solute carrier family 17 member 1) (Renal sodium-dependent phosphate transport protein 1) (Renal sodium-phosphate transport protein 1) (Renal Na(+)-dependent phosphate cotransporter 1) (Na/Pi-4). | |||||
|
ELOV4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 11) | NC score | 0.042695 (rank : 8) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQC4 | Gene names | Elovl4 | |||
|
Domain Architecture |
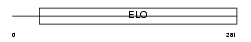
|
|||||
| Description | Elongation of very long chain fatty acids protein 4. | |||||
|
CTL2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 12) | NC score | 0.025971 (rank : 11) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BY89, Q8K2F1 | Gene names | Slc44a2, Ctl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Choline transporter-like protein 2 (Solute carrier family 44 member 2). | |||||
|
CTL2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 13) | NC score | 0.017023 (rank : 14) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWA5, Q658V1, Q658Z2, Q6PJV7, Q8N2F0, Q9NY68 | Gene names | SLC44A2, CTL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Choline transporter-like protein 2 (Solute carrier family 44 member 2). | |||||
|
PARG_HUMAN
|
||||||
| θ value | 6.88961 (rank : 14) | NC score | 0.035439 (rank : 10) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86W56, Q6E4P6, Q6E4P7, Q7Z742, Q9Y4W7 | Gene names | PARG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
PROM1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 15) | NC score | 0.013984 (rank : 15) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43490 | Gene names | PROM1, PROML1 | |||
|
Domain Architecture |

|
|||||
| Description | Prominin-1 precursor (Prominin-like protein 1) (Antigen AC133) (CD133 antigen). | |||||
|
ELTD1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 16) | NC score | 0.005231 (rank : 18) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBW9 | Gene names | ELTD1, ETL | |||
|
Domain Architecture |

|
|||||
| Description | EGF, latrophilin and seven transmembrane domain-containing protein 1 precursor (EGF-TM7-latrophilin-related protein) (ETL protein). | |||||
|
ELTD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.004396 (rank : 19) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q923X1, Q91W44 | Gene names | Eltd1 | |||
|
Domain Architecture |

|
|||||
| Description | EGF, latrophilin seven transmembrane domain-containing protein 1 precursor. | |||||
|
MNT_MOUSE
|
||||||
| θ value | 8.99809 (rank : 18) | NC score | 0.007917 (rank : 16) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
SL9A1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | 0.006805 (rank : 17) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61165 | Gene names | Slc9a1, Nhe1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/hydrogen exchanger 1 (Na(+)/H(+) exchanger 1) (NHE-1) (Solute carrier family 9 member 1). | |||||
|
CDS2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99L43, Q3TMD1, Q6NSU1 | Gene names | Cds2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidate cytidylyltransferase 2 (EC 2.7.7.41) (CDP-diglyceride synthetase 2) (CDP-diglyceride pyrophosphorylase 2) (CDP- diacylglycerol synthase 2) (CDS 2) (CTP:phosphatidate cytidylyltransferase 2) (CDP-DAG synthase 2) (CDP-DG synthetase 2). | |||||
|
CDS2_HUMAN
|
||||||
| NC score | 0.998698 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O95674, Q5TDY2, Q5TDY3, Q5TDY4, Q5TDY5, Q9BYK5, Q9NTT2 | Gene names | CDS2 | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidate cytidylyltransferase 2 (EC 2.7.7.41) (CDP-diglyceride synthetase 2) (CDP-diglyceride pyrophosphorylase 2) (CDP- diacylglycerol synthase 2) (CDS 2) (CTP:phosphatidate cytidylyltransferase 2) (CDP-DAG synthase 2) (CDP-DG synthetase 2). | |||||
|
CDS1_MOUSE
|
||||||
| NC score | 0.994179 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P98191, Q8CGZ1 | Gene names | Cds1, Cds | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Phosphatidate cytidylyltransferase 1 (EC 2.7.7.41) (CDP-diglyceride synthetase 1) (CDP-diglyceride pyrophosphorylase 1) (CDP- diacylglycerol synthase 1) (CDS 1) (CTP:phosphatidate cytidylyltransferase 1) (CDP-DAG synthase 1) (CDP-DG synthetase 1). | |||||
|
CDS1_HUMAN
|
||||||
| NC score | 0.993455 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92903, O00163 | Gene names | CDS1, CDS | |||
|
Domain Architecture |

|
|||||
| Description | Phosphatidate cytidylyltransferase 1 (EC 2.7.7.41) (CDP-diglyceride synthetase 1) (CDP-diglyceride pyrophosphorylase 1) (CDP- diacylglycerol synthase 1) (CDS 1) (CTP:phosphatidate cytidylyltransferase 1) (CDP-DAG synthase 1) (CDP-DG synthetase 1). | |||||
|
ELOV4_HUMAN
|
||||||
| NC score | 0.064467 (rank : 5) | θ value | 0.163984 (rank : 7) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZR5, Q5TCS2, Q86YJ1, Q9H139 | Gene names | ELOVL4 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation of very long chain fatty acids protein 4. | |||||
|
PARG_MOUSE
|
||||||
| NC score | 0.057376 (rank : 6) | θ value | 0.21417 (rank : 8) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 54 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88622, Q80YQ6, Q8CB72 | Gene names | Parg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
G109B_HUMAN
|
||||||
| NC score | 0.042703 (rank : 7) | θ value | 0.0563607 (rank : 6) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 579 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P49019, Q8NGE4 | Gene names | GPR109B | |||
|
Domain Architecture |

|
|||||
| Description | Nicotinic acid receptor 2 (G-protein coupled receptor 109B) (G-protein coupled receptor HM74) (G-protein coupled receptor HM74B). | |||||
|
ELOV4_MOUSE
|
||||||
| NC score | 0.042695 (rank : 8) | θ value | 4.03905 (rank : 11) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9EQC4 | Gene names | Elovl4 | |||
|
Domain Architecture |

|
|||||
| Description | Elongation of very long chain fatty acids protein 4. | |||||
|
G109A_HUMAN
|
||||||
| NC score | 0.039297 (rank : 9) | θ value | 0.0563607 (rank : 5) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 614 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8TDS4 | Gene names | GPR109A, HM74A | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nicotinic acid receptor 1 (G-protein coupled receptor 109A) (G-protein coupled receptor HM74A). | |||||
|
PARG_HUMAN
|
||||||
| NC score | 0.035439 (rank : 10) | θ value | 6.88961 (rank : 14) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 97 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86W56, Q6E4P6, Q6E4P7, Q7Z742, Q9Y4W7 | Gene names | PARG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Poly(ADP-ribose) glycohydrolase (EC 3.2.1.143). | |||||
|
CTL2_MOUSE
|
||||||
| NC score | 0.025971 (rank : 11) | θ value | 5.27518 (rank : 12) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BY89, Q8K2F1 | Gene names | Slc44a2, Ctl2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Choline transporter-like protein 2 (Solute carrier family 44 member 2). | |||||
|
NPT1_HUMAN
|
||||||
| NC score | 0.021751 (rank : 12) | θ value | 3.0926 (rank : 10) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14916, Q13783 | Gene names | SLC17A1, NPT1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium-dependent phosphate transport protein 1 (Sodium/phosphate cotransporter 1) (Na(+)/PI cotransporter 1) (Solute carrier family 17 member 1) (Renal sodium-dependent phosphate transport protein 1) (Renal sodium-phosphate transport protein 1) (Renal Na(+)-dependent phosphate cotransporter 1) (Na/Pi-4). | |||||
|
GP109_MOUSE
|
||||||
| NC score | 0.020814 (rank : 13) | θ value | 2.36792 (rank : 9) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 588 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9EP66 | Gene names | Gpr109, Gpr109a, Gpr109b, Pumag | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nicotinic acid receptor (G-protein coupled receptor 109) (G-protein coupled receptor HM74) (Protein PUMA-G). | |||||
|
CTL2_HUMAN
|
||||||
| NC score | 0.017023 (rank : 14) | θ value | 6.88961 (rank : 13) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IWA5, Q658V1, Q658Z2, Q6PJV7, Q8N2F0, Q9NY68 | Gene names | SLC44A2, CTL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Choline transporter-like protein 2 (Solute carrier family 44 member 2). | |||||
|
PROM1_HUMAN
|
||||||
| NC score | 0.013984 (rank : 15) | θ value | 6.88961 (rank : 15) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O43490 | Gene names | PROM1, PROML1 | |||
|
Domain Architecture |

|
|||||
| Description | Prominin-1 precursor (Prominin-like protein 1) (Antigen AC133) (CD133 antigen). | |||||
|
MNT_MOUSE
|
||||||
| NC score | 0.007917 (rank : 16) | θ value | 8.99809 (rank : 18) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O08789, P97349 | Gene names | Mnt, Rox | |||
|
Domain Architecture |

|
|||||
| Description | Max-binding protein MNT (Protein ROX) (Myc antagonist MNT). | |||||
|
SL9A1_MOUSE
|
||||||
| NC score | 0.006805 (rank : 17) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q61165 | Gene names | Slc9a1, Nhe1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/hydrogen exchanger 1 (Na(+)/H(+) exchanger 1) (NHE-1) (Solute carrier family 9 member 1). | |||||
|
ELTD1_HUMAN
|
||||||
| NC score | 0.005231 (rank : 18) | θ value | 8.99809 (rank : 16) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 269 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9HBW9 | Gene names | ELTD1, ETL | |||
|
Domain Architecture |

|
|||||
| Description | EGF, latrophilin and seven transmembrane domain-containing protein 1 precursor (EGF-TM7-latrophilin-related protein) (ETL protein). | |||||
|
ELTD1_MOUSE
|
||||||
| NC score | 0.004396 (rank : 19) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 19 | Target Neighborhood Hits | 288 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q923X1, Q91W44 | Gene names | Eltd1 | |||
|
Domain Architecture |

|
|||||
| Description | EGF, latrophilin seven transmembrane domain-containing protein 1 precursor. | |||||