Please be patient as the page loads
|
CCDC8_HUMAN
|
||||||
| SwissProt Accessions | Q9H0W5, Q8TB26 | Gene names | CCDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 8. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
CCDC8_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9H0W5, Q8TB26 | Gene names | CCDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 8. | |||||
|
PNMA1_HUMAN
|
||||||
| θ value | 7.09661e-13 (rank : 2) | NC score | 0.500010 (rank : 2) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8ND90, O95144, Q8NG07 | Gene names | PNMA1, MA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paraneoplastic antigen Ma1 (Neuron- and testis-specific protein 1) (37 kDa neuronal protein). | |||||
|
PNMA1_MOUSE
|
||||||
| θ value | 4.30538e-10 (rank : 3) | NC score | 0.488012 (rank : 3) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C1C8, Q9CYP2 | Gene names | Pnma1, Ma1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paraneoplastic antigen Ma1 homolog. | |||||
|
RRBP1_HUMAN
|
||||||
| θ value | 4.30538e-10 (rank : 4) | NC score | 0.170677 (rank : 12) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
PNMA2_MOUSE
|
||||||
| θ value | 6.21693e-09 (rank : 5) | NC score | 0.476993 (rank : 4) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BHK0 | Gene names | Pnma2 | |||
|
Domain Architecture |

|
|||||
| Description | Paraneoplastic antigen Ma2 homolog. | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 3.08544e-08 (rank : 6) | NC score | 0.161992 (rank : 16) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
PNMA2_HUMAN
|
||||||
| θ value | 8.97725e-08 (rank : 7) | NC score | 0.466401 (rank : 5) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UL42, O94959, O95145, Q9UL43 | Gene names | PNMA2, KIAA0883, MA2 | |||
|
Domain Architecture |

|
|||||
| Description | Paraneoplastic antigen Ma2 (Onconeuronal antigen MA2) (Paraneoplastic neuronal antigen MM2) (40 kDa neuronal protein). | |||||
|
MOAP1_MOUSE
|
||||||
| θ value | 3.77169e-06 (rank : 8) | NC score | 0.456357 (rank : 6) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ERH6 | Gene names | Moap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Modulator of apoptosis 1 (MAP-1). | |||||
|
CLIC6_HUMAN
|
||||||
| θ value | 1.09739e-05 (rank : 9) | NC score | 0.179960 (rank : 11) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
MOAP1_HUMAN
|
||||||
| θ value | 4.1701e-05 (rank : 10) | NC score | 0.449527 (rank : 7) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96BY2, Q9H833, Q9HAS1 | Gene names | MOAP1, PNMA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Modulator of apoptosis 1 (MAP-1) (MAP1) (Paraneoplastic antigen MA4). | |||||
|
IWS1_HUMAN
|
||||||
| θ value | 7.1131e-05 (rank : 11) | NC score | 0.169733 (rank : 13) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| θ value | 0.000786445 (rank : 12) | NC score | 0.166763 (rank : 14) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
LEO1_HUMAN
|
||||||
| θ value | 0.00390308 (rank : 13) | NC score | 0.158505 (rank : 20) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 14) | NC score | 0.067006 (rank : 100) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
FILA_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 15) | NC score | 0.162636 (rank : 15) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
IWS1_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 16) | NC score | 0.147394 (rank : 21) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
MARCS_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 17) | NC score | 0.213187 (rank : 10) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
RPTN_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 18) | NC score | 0.130227 (rank : 31) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
AP3B2_MOUSE
|
||||||
| θ value | 0.163984 (rank : 19) | NC score | 0.087724 (rank : 60) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
TARA_HUMAN
|
||||||
| θ value | 0.163984 (rank : 20) | NC score | 0.077651 (rank : 74) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
NALP5_MOUSE
|
||||||
| θ value | 0.21417 (rank : 21) | NC score | 0.057437 (rank : 124) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
ZBT17_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.010792 (rank : 172) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
TB182_HUMAN
|
||||||
| θ value | 0.279714 (rank : 23) | NC score | 0.118479 (rank : 36) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
ZKSC1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 24) | NC score | 0.011063 (rank : 171) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17029, P52745, Q8TBW5, Q8TEK7 | Gene names | ZKSCAN1, KOX18, ZNF139, ZNF36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger with KRAB and SCAN domain-containing protein 1 (Zinc finger protein 36) (Zinc finger protein KOX18). | |||||
|
DSPP_HUMAN
|
||||||
| θ value | 0.47712 (rank : 25) | NC score | 0.159492 (rank : 17) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
JARD2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 26) | NC score | 0.070184 (rank : 96) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
PEG3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 27) | NC score | 0.018203 (rank : 169) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9GZU2, P78418, Q5H9P9, Q7Z7H7, Q8TF75, Q9GZY2 | Gene names | PEG3, KIAA0287 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein. | |||||
|
SRCA_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.131120 (rank : 28) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
AKA12_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.107257 (rank : 45) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
CAMKV_HUMAN
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.008889 (rank : 176) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
TARA_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.074383 (rank : 85) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.132383 (rank : 26) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
SCG1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 33) | NC score | 0.130568 (rank : 29) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P16014 | Gene names | Chgb, Scg-1, Scg1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB). | |||||
|
SPTB1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.024257 (rank : 162) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
TRI44_HUMAN
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.066242 (rank : 101) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
ZC313_HUMAN
|
||||||
| θ value | 1.81305 (rank : 36) | NC score | 0.116982 (rank : 38) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
MDN1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.115705 (rank : 39) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
PEG3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.031662 (rank : 159) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
ABL1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.004586 (rank : 178) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
CLIC6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.087828 (rank : 57) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
DAXX_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.087822 (rank : 58) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
DIDO1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.048075 (rank : 152) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
MYST4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.046624 (rank : 153) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
B3A3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.020947 (rank : 165) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48751 | Gene names | SLC4A3, AE3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3) (Cardiac/brain band 3-like protein) (CAE3/BAE3). | |||||
|
CJ086_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.045649 (rank : 154) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXX6, Q5SQQ5, Q6P673, Q8WY66, Q9BS90 | Gene names | C10orf86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf86. | |||||
|
CLCB_HUMAN
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.031388 (rank : 160) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09497, Q6FHW1 | Gene names | CLTB | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
DDX55_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.009146 (rank : 175) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.023266 (rank : 163) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
VGF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.125754 (rank : 35) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15240 | Gene names | VGF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurosecretory protein VGF precursor. | |||||
|
ZAR1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.063055 (rank : 109) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
ZAR1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.065188 (rank : 105) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
BRD4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.037573 (rank : 157) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
FGD4_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.009863 (rank : 173) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
K22O_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.013867 (rank : 170) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01546, Q7Z795 | Gene names | KRT76, KRT2B, KRT2P | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 oral (Cytokeratin-2P) (K2P) (CK 2P) (Keratin 76). | |||||
|
MINT_MOUSE
|
||||||
| θ value | 5.27518 (rank : 55) | NC score | 0.058766 (rank : 118) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
PARPT_HUMAN
|
||||||
| θ value | 5.27518 (rank : 56) | NC score | 0.018840 (rank : 168) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
RBM25_HUMAN
|
||||||
| θ value | 5.27518 (rank : 57) | NC score | 0.041805 (rank : 155) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
B3A3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.020332 (rank : 166) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16283 | Gene names | Slc4a3, Ae3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3). | |||||
|
BTBDC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.041284 (rank : 156) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
CORIN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.002457 (rank : 179) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
MRP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.113274 (rank : 43) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
PESC_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.031912 (rank : 158) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00541, Q6IC29 | Gene names | PES1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
SMC4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.019949 (rank : 167) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
SPTB1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.022449 (rank : 164) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
ZBT17_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.009599 (rank : 174) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |
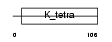
|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||
|
AN32B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.052734 (rank : 139) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
HIRP3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.131791 (rank : 27) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
LRC45_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.029381 (rank : 161) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
MARCS_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.159286 (rank : 19) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
PTN18_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.005666 (rank : 177) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
ACINU_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.056736 (rank : 127) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
AFF4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.057364 (rank : 125) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.065920 (rank : 103) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
ALO17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.059522 (rank : 115) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
AP3B1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.070822 (rank : 94) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.058187 (rank : 121) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
AP3B2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.056778 (rank : 126) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
ARS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.067668 (rank : 99) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
ARS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.077304 (rank : 76) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
ASPX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.091040 (rank : 54) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
ATRX_HUMAN
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.054986 (rank : 132) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
ATRX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.055043 (rank : 131) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
BASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.127501 (rank : 33) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
BASP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.133943 (rank : 25) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
CCD55_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.077550 (rank : 75) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
CENPB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.060255 (rank : 114) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
CF010_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.087767 (rank : 59) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
CIR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.063413 (rank : 108) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
CMGA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.069540 (rank : 98) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
CTR9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.071297 (rank : 92) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
CTR9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.075776 (rank : 82) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
CYLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.086103 (rank : 62) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
CYLC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.134523 (rank : 24) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
DAXX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.062056 (rank : 110) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
DMP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.130193 (rank : 32) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
DMP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.138210 (rank : 23) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.159410 (rank : 18) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ENL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.071833 (rank : 89) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.059368 (rank : 116) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
FUS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.058298 (rank : 119) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
FUS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.059191 (rank : 117) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
GNAS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.054701 (rank : 133) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
GPTC8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.113926 (rank : 42) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
H13_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.053492 (rank : 136) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
H14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.050021 (rank : 150) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
HORN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.115486 (rank : 40) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
HORN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.117134 (rank : 37) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
HTSF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.079682 (rank : 69) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
HTSF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.081190 (rank : 66) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
IF2P_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.051118 (rank : 144) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
LRRF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.076958 (rank : 77) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
MAP1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.055359 (rank : 130) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
MRP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.115253 (rank : 41) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
NASP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.071714 (rank : 91) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
NCKX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.076071 (rank : 81) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
NEUM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.085260 (rank : 63) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
NEUM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.053060 (rank : 138) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
NFH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.051104 (rank : 145) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
NIPBL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.074814 (rank : 83) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
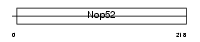
NNP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.061349 (rank : 113) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P56183, O35712, Q9ERE1, Q9JI07, Q9JK67, Q9JKU2 | Gene names | Nnp1 | |||
|
Domain Architecture |
|
|||||
| Description | NNP-1 protein (Novel nuclear protein 1) (Nop52). | |||||
|
NOLC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.096988 (rank : 52) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
NPBL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.077715 (rank : 73) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
NSBP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.145927 (rank : 22) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
NSBP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.125906 (rank : 34) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
NUCKS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.109481 (rank : 44) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
NUCKS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.106469 (rank : 46) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
PPIG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.050754 (rank : 148) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.063481 (rank : 107) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
PPRB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.058193 (rank : 120) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
PRB3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.101317 (rank : 48) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
PRB4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.070488 (rank : 95) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.105787 (rank : 47) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRP5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.054508 (rank : 135) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
PRPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.073726 (rank : 86) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
PRPE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.053141 (rank : 137) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
PTMA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.095251 (rank : 53) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
PTMA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.099405 (rank : 51) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
PTMS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.057483 (rank : 123) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
PTMS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.052039 (rank : 143) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.056480 (rank : 128) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
RBBP6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.071806 (rank : 90) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
RPTN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.082486 (rank : 65) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
SCG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.089964 (rank : 55) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
SEMG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.065925 (rank : 102) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
SFR11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.076492 (rank : 78) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
SFR12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.081121 (rank : 67) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
SFR12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.087889 (rank : 56) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
SFR17_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.085228 (rank : 64) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.052620 (rank : 140) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
SFRS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.078087 (rank : 72) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.072620 (rank : 88) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
SH3BG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.051001 (rank : 146) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WUZ7 | Gene names | Sh3bgr | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding glutamic acid-rich protein (SH3BGR protein). | |||||
|
SIAL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.076164 (rank : 80) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P21815 | Gene names | IBSP, BNSP | |||
|
Domain Architecture |
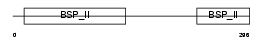
|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
SPRL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.076334 (rank : 79) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
SRCA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.101001 (rank : 49) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
SRCH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.078111 (rank : 71) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
SRRM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.063817 (rank : 106) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
SRRM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.052536 (rank : 142) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
T2FA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.100772 (rank : 50) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
T2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.130402 (rank : 30) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06666 | Gene names | Srst, T2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Octapeptide-repeat protein T2. | |||||
|
TCAL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.050919 (rank : 147) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H3H9 | Gene names | TCEAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 2 (TCEA-like protein 2) (Transcription elongation factor S-II protein-like 2). | |||||
|
TCAL3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.061621 (rank : 112) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R0A5, Q9CYK5, Q9D1S4 | Gene names | Tceal3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
TCAL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.061669 (rank : 111) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TCAL5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.054585 (rank : 134) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CCT4, Q497R6 | Gene names | Tceal5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.073084 (rank : 87) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TCOF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.070176 (rank : 97) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
TGON2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.087570 (rank : 61) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
THOC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.055573 (rank : 129) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
TNC6B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.057697 (rank : 122) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
TR150_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.050020 (rank : 151) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
TRDN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.078707 (rank : 70) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
TXND2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.052571 (rank : 141) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.074527 (rank : 84) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
VCX3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.065598 (rank : 104) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
VCXC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.070980 (rank : 93) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
YTDC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.081086 (rank : 68) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
ZCH10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.050203 (rank : 149) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
ZCH12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.274366 (rank : 9) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PEW1, Q6PID5, Q8N1C1 | Gene names | ZCCHC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 12. | |||||
|
ZCH12_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.275087 (rank : 8) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CZA5 | Gene names | Zcchc12, Sizn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 12 (Smad-interacting zinc finger protein). | |||||
|
CCDC8_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 70 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q9H0W5, Q8TB26 | Gene names | CCDC8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 8. | |||||
|
PNMA1_HUMAN
|
||||||
| NC score | 0.500010 (rank : 2) | θ value | 7.09661e-13 (rank : 2) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8ND90, O95144, Q8NG07 | Gene names | PNMA1, MA1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paraneoplastic antigen Ma1 (Neuron- and testis-specific protein 1) (37 kDa neuronal protein). | |||||
|
PNMA1_MOUSE
|
||||||
| NC score | 0.488012 (rank : 3) | θ value | 4.30538e-10 (rank : 3) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8C1C8, Q9CYP2 | Gene names | Pnma1, Ma1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paraneoplastic antigen Ma1 homolog. | |||||
|
PNMA2_MOUSE
|
||||||
| NC score | 0.476993 (rank : 4) | θ value | 6.21693e-09 (rank : 5) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BHK0 | Gene names | Pnma2 | |||
|
Domain Architecture |

|
|||||
| Description | Paraneoplastic antigen Ma2 homolog. | |||||
|
PNMA2_HUMAN
|
||||||
| NC score | 0.466401 (rank : 5) | θ value | 8.97725e-08 (rank : 7) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UL42, O94959, O95145, Q9UL43 | Gene names | PNMA2, KIAA0883, MA2 | |||
|
Domain Architecture |

|
|||||
| Description | Paraneoplastic antigen Ma2 (Onconeuronal antigen MA2) (Paraneoplastic neuronal antigen MM2) (40 kDa neuronal protein). | |||||
|
MOAP1_MOUSE
|
||||||
| NC score | 0.456357 (rank : 6) | θ value | 3.77169e-06 (rank : 8) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ERH6 | Gene names | Moap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Modulator of apoptosis 1 (MAP-1). | |||||
|
MOAP1_HUMAN
|
||||||
| NC score | 0.449527 (rank : 7) | θ value | 4.1701e-05 (rank : 10) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q96BY2, Q9H833, Q9HAS1 | Gene names | MOAP1, PNMA4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Modulator of apoptosis 1 (MAP-1) (MAP1) (Paraneoplastic antigen MA4). | |||||
|
ZCH12_MOUSE
|
||||||
| NC score | 0.275087 (rank : 8) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CZA5 | Gene names | Zcchc12, Sizn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 12 (Smad-interacting zinc finger protein). | |||||
|
ZCH12_HUMAN
|
||||||
| NC score | 0.274366 (rank : 9) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6PEW1, Q6PID5, Q8N1C1 | Gene names | ZCCHC12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 12. | |||||
|
MARCS_MOUSE
|
||||||
| NC score | 0.213187 (rank : 10) | θ value | 0.0563607 (rank : 17) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | P26645 | Gene names | Marcks, Macs | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS). | |||||
|
CLIC6_HUMAN
|
||||||
| NC score | 0.179960 (rank : 11) | θ value | 1.09739e-05 (rank : 9) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q96NY7, Q8IX31 | Gene names | CLIC6, CLIC1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
RRBP1_HUMAN
|
||||||
| NC score | 0.170677 (rank : 12) | θ value | 4.30538e-10 (rank : 4) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1306 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9P2E9, O75300, O75301, Q5W165, Q96SB2, Q9BWP1, Q9H476 | Gene names | RRBP1, KIAA1398 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (180 kDa ribosome receptor homolog) (ES/130-related protein). | |||||
|
IWS1_HUMAN
|
||||||
| NC score | 0.169733 (rank : 13) | θ value | 7.1131e-05 (rank : 11) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1241 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q96ST2, Q6P157, Q8N3E8, Q96MI7, Q9NV97, Q9NWH8 | Gene names | IWS1, IWS1L | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
LEO1_MOUSE
|
||||||
| NC score | 0.166763 (rank : 14) | θ value | 0.000786445 (rank : 12) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 946 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q5XJE5, Q640R1 | Gene names | Leo1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1. | |||||
|
FILA_HUMAN
|
||||||
| NC score | 0.162636 (rank : 15) | θ value | 0.0330416 (rank : 15) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P20930, Q01720, Q5T583 | Gene names | FLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filaggrin. | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.161992 (rank : 16) | θ value | 3.08544e-08 (rank : 6) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
DSPP_HUMAN
|
||||||
| NC score | 0.159492 (rank : 17) | θ value | 0.47712 (rank : 25) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 645 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9NZW4, O95815 | Gene names | DSPP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.159410 (rank : 18) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
MARCS_HUMAN
|
||||||
| NC score | 0.159286 (rank : 19) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P29966, Q2LA83, Q5TDB7 | Gene names | MARCKS, MACS, PRKCSL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate, 80 kDa protein, light chain) (PKCSL) (80K-L protein). | |||||
|
LEO1_HUMAN
|
||||||
| NC score | 0.158505 (rank : 20) | θ value | 0.00390308 (rank : 13) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q8WVC0, Q96N99 | Gene names | LEO1, RDL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein LEO1 (Replicative senescence down- regulated leo1-like protein). | |||||
|
IWS1_MOUSE
|
||||||
| NC score | 0.147394 (rank : 21) | θ value | 0.0563607 (rank : 16) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1258 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8C1D8, Q3TQ25, Q3UWB0, Q6NVD1, Q8BY73, Q9D9H4 | Gene names | Iws1, Iws1l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | IWS1 homolog (IWS1-like protein). | |||||
|
NSBP1_HUMAN
|
||||||
| NC score | 0.145927 (rank : 22) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P82970 | Gene names | NSBP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1. | |||||
|
DMP1_MOUSE
|
||||||
| NC score | 0.138210 (rank : 23) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O55188 | Gene names | Dmp1, Dmp | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1) (AG1). | |||||
|
CYLC2_HUMAN
|
||||||
| NC score | 0.134523 (rank : 24) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q14093 | Gene names | CYLC2, CYL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-2 (Cylicin II) (Multiple-band polypeptide II). | |||||
|
BASP_MOUSE
|
||||||
| NC score | 0.133943 (rank : 25) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 260 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q91XV3 | Gene names | Basp1, Nap22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.132383 (rank : 26) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
HIRP3_HUMAN
|
||||||
| NC score | 0.131791 (rank : 27) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BW71, O75707, O75708 | Gene names | HIRIP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIRA-interacting protein 3. | |||||
|
SRCA_HUMAN
|
||||||
| NC score | 0.131120 (rank : 28) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q86TD4 | Gene names | SRL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
SCG1_MOUSE
|
||||||
| NC score | 0.130568 (rank : 29) | θ value | 1.38821 (rank : 33) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P16014 | Gene names | Chgb, Scg-1, Scg1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB). | |||||
|
T2_MOUSE
|
||||||
| NC score | 0.130402 (rank : 30) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q06666 | Gene names | Srst, T2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Octapeptide-repeat protein T2. | |||||
|
RPTN_MOUSE
|
||||||
| NC score | 0.130227 (rank : 31) | θ value | 0.0736092 (rank : 18) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P97347 | Gene names | Rptn | |||
|
Domain Architecture |

|
|||||
| Description | Repetin. | |||||
|
DMP1_HUMAN
|
||||||
| NC score | 0.130193 (rank : 32) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13316, O43265 | Gene names | DMP1 | |||
|
Domain Architecture |

|
|||||
| Description | Dentin matrix acidic phosphoprotein 1 precursor (Dentin matrix protein 1) (DMP-1). | |||||
|
BASP_HUMAN
|
||||||
| NC score | 0.127501 (rank : 33) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 321 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P80723, O43596, Q5U0S0 | Gene names | BASP1, NAP22 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain acid soluble protein 1 (BASP1 protein) (Neuronal axonal membrane protein NAP-22) (22 kDa neuronal tissue-enriched acidic protein). | |||||
|
NSBP1_MOUSE
|
||||||
| NC score | 0.125906 (rank : 34) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 681 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9JL35, O88832, Q8VC71, Q9CUW1 | Gene names | Nsbp1, Garp45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleosome-binding protein 1 (Nucleosome-binding protein 45) (NBP-45) (GARP45 protein). | |||||
|
VGF_HUMAN
|
||||||
| NC score | 0.125754 (rank : 35) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O15240 | Gene names | VGF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurosecretory protein VGF precursor. | |||||
|
TB182_HUMAN
|
||||||
| NC score | 0.118479 (rank : 36) | θ value | 0.279714 (rank : 23) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 414 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9C0C2 | Gene names | TNKS1BP1, KIAA1741, TAB182 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 182 kDa tankyrase 1-binding protein. | |||||
|
HORN_MOUSE
|
||||||
| NC score | 0.117134 (rank : 37) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8VHD8 | Gene names | Hrnr | |||
|
Domain Architecture |

|
|||||
| Description | Hornerin. | |||||
|
ZC313_HUMAN
|
||||||
| NC score | 0.116982 (rank : 38) | θ value | 1.81305 (rank : 36) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 772 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q5T200, O94936, Q5T1Z9, Q7Z7J3, Q8NDT6, Q9H0L6 | Gene names | ZC3H13, KIAA0853 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein 13. | |||||
|
MDN1_HUMAN
|
||||||
| NC score | 0.115705 (rank : 39) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 662 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9NU22, O15019 | Gene names | MDN1, KIAA0301 | |||
|
Domain Architecture |

|
|||||
| Description | Midasin (MIDAS-containing protein). | |||||
|
HORN_HUMAN
|
||||||
| NC score | 0.115486 (rank : 40) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 206 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q86YZ3 | Gene names | HRNR, S100A18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hornerin. | |||||
|
MRP_HUMAN
|
||||||
| NC score | 0.115253 (rank : 41) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 51 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P49006, Q5TEE6, Q6NXS5 | Gene names | MARCKSL1, MLP, MRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS). | |||||
|
GPTC8_HUMAN
|
||||||
| NC score | 0.113926 (rank : 42) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 366 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9UKJ3, O60300 | Gene names | GPATC8, KIAA0553 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | G patch domain-containing protein 8. | |||||
|
MRP_MOUSE
|
||||||
| NC score | 0.113274 (rank : 43) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P28667, Q3TEZ4, Q91W07 | Gene names | Marcksl1, Mlp, Mrp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MARCKS-related protein (MARCKS-like protein 1) (Macrophage myristoylated alanine-rich C kinase substrate) (Mac-MARCKS) (MacMARCKS) (Brain protein F52). | |||||
|
NUCKS_HUMAN
|
||||||
| NC score | 0.109481 (rank : 44) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9H1E3, Q9H1D6, Q9H723 | Gene names | NUCKS1, NUCKS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (P1). | |||||
|
AKA12_HUMAN
|
||||||
| NC score | 0.107257 (rank : 45) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 825 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q02952, O00310, O00498, Q99970 | Gene names | AKAP12, AKAP250 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | A-kinase anchor protein 12 (A-kinase anchor protein 250 kDa) (AKAP 250) (Myasthenia gravis autoantigen gravin). | |||||
|
NUCKS_MOUSE
|
||||||
| NC score | 0.106469 (rank : 46) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 114 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80XU3, Q8BVD8 | Gene names | Nucks1, Nucks | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear ubiquitous casein and cyclin-dependent kinases substrate (JC7). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.105787 (rank : 47) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
PRB3_HUMAN
|
||||||
| NC score | 0.101317 (rank : 48) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q04118, Q15188, Q7M4M9 | Gene names | PRB3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 3 precursor (Parotid salivary glycoprotein G1) (Proline-rich protein G1). | |||||
|
SRCA_MOUSE
|
||||||
| NC score | 0.101001 (rank : 49) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q7TQ48, Q8BS43 | Gene names | Srl, Sar | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcalumenin precursor. | |||||
|
T2FA_HUMAN
|
||||||
| NC score | 0.100772 (rank : 50) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 228 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35269, Q9BWN0 | Gene names | GTF2F1, RAP74 | |||
|
Domain Architecture |

|
|||||
| Description | Transcription initiation factor IIF alpha subunit (EC 2.7.11.1) (TFIIF-alpha) (Transcription initiation factor RAP74) (General transcription factor IIF polypeptide 1 74 kDa subunit protein). | |||||
|
PTMA_MOUSE
|
||||||
| NC score | 0.099405 (rank : 51) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P26350, Q3UQV6 | Gene names | Ptma | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha]. | |||||
|
NOLC1_HUMAN
|
||||||
| NC score | 0.096988 (rank : 52) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 452 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q14978, Q15030 | Gene names | NOLC1, KIAA0035 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar phosphoprotein p130 (Nucleolar 130 kDa protein) (140 kDa nucleolar phosphoprotein) (Nopp140) (Nucleolar and coiled-body phosphoprotein 1). | |||||
|
PTMA_HUMAN
|
||||||
| NC score | 0.095251 (rank : 53) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 88 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P06454, Q15249, Q15592 | Gene names | PTMA, TMSA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Prothymosin alpha [Contains: Thymosin alpha-1]. | |||||
|
ASPX_HUMAN
|
||||||
| NC score | 0.091040 (rank : 54) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P26436 | Gene names | ACRV1 | |||
|
Domain Architecture |

|
|||||
| Description | Acrosomal protein SP-10 precursor (Acrosomal vesicle protein 1). | |||||
|
SCG1_HUMAN
|
||||||
| NC score | 0.089964 (rank : 55) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P05060, Q9BQV6, Q9UJA6 | Gene names | CHGB, SCG1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB) [Contains: GAWK peptide; CCB peptide]. | |||||
|
SFR12_MOUSE
|
||||||
| NC score | 0.087889 (rank : 56) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 486 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q8BZX4, Q8BLM8, Q8BWK7, Q8BYK0, Q8BYZ4 | Gene names | Sfrs12, Srrp86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86). | |||||
|
CLIC6_MOUSE
|
||||||
| NC score | 0.087828 (rank : 57) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8BHB9 | Gene names | Clic6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chloride intracellular channel 6. | |||||
|
DAXX_HUMAN
|
||||||
| NC score | 0.087822 (rank : 58) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 207 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9UER7, O14747, O15141, O15208, Q9BWI3 | Gene names | DAXX, BING2, DAP6 | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx) (hDaxx) (Fas death domain- associated protein) (ETS1-associated protein 1) (EAP1). | |||||
|
CF010_HUMAN
|
||||||
| NC score | 0.087767 (rank : 59) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 329 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q5SRN2, Q5SPI9, Q5SPJ0, Q5SPK9, Q5SPL0, Q5SRN3, Q5TG25, Q5TG26, Q8N4B6 | Gene names | C6orf10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf10. | |||||
|
AP3B2_MOUSE
|
||||||
| NC score | 0.087724 (rank : 60) | θ value | 0.163984 (rank : 19) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JME5, Q6QR53, Q8R1E5 | Gene names | Ap3b2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain). | |||||
|
TGON2_HUMAN
|
||||||
| NC score | 0.087570 (rank : 61) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1037 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | O43493, O15282, O43492, O43499, O43500, O43501, Q92760 | Gene names | TGOLN2, TGN46, TGN51 | |||
|
Domain Architecture |

|
|||||
| Description | Trans-Golgi network integral membrane protein 2 precursor (Trans-Golgi network protein TGN51) (TGN46) (TGN48) (TGN38 homolog). | |||||
|
CYLC1_HUMAN
|
||||||
| NC score | 0.086103 (rank : 62) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P35663, Q5JQQ9 | Gene names | CYLC1, CYL, CYL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cylicin-1 (Cylicin I) (Multiple-band polypeptide I). | |||||
|
NEUM_HUMAN
|
||||||
| NC score | 0.085260 (rank : 63) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P17677 | Gene names | GAP43 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (PP46) (Neural phosphoprotein B-50). | |||||
|
SFR17_HUMAN
|
||||||
| NC score | 0.085228 (rank : 64) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 325 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8TF01, Q5T064, Q6P2B4, Q6P2N4, Q6PJ93, Q6PK36, Q7Z640, Q8TF00, Q96K10, Q96SM5, Q9P076, Q9P0C0, Q9Y4N3 | Gene names | SRRP130, C6orf111 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, arginine/serine-rich 130 (Serine-arginine-rich- splicing regulatory protein 130) (SRrp130) (SR-rich protein) (SR- related protein). | |||||
|
RPTN_HUMAN
|
||||||
| NC score | 0.082486 (rank : 65) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q6XPR3 | Gene names | RPTN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Repetin. | |||||
|
HTSF1_MOUSE
|
||||||
| NC score | 0.081190 (rank : 66) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q8BGC0, Q1WWK0, Q9CT41, Q9DAU3 | Gene names | Htatsf1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 homolog. | |||||
|
SFR12_HUMAN
|
||||||
| NC score | 0.081121 (rank : 67) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8WXA9, Q86X37 | Gene names | SFRS12, SRRP86 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 12 (Serine-arginine-rich- splicing regulatory protein 86) (SRrp86) (Splicing regulatory protein 508) (SRrp508). | |||||
|
YTDC1_HUMAN
|
||||||
| NC score | 0.081086 (rank : 68) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 458 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q96MU7, Q7Z622, Q8TF35 | Gene names | YTHDC1, KIAA1966, YT521 | |||
|
Domain Architecture |

|
|||||
| Description | YTH domain-containing protein 1 (Putative splicing factor YT521). | |||||
|
HTSF1_HUMAN
|
||||||
| NC score | 0.079682 (rank : 69) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O43719, Q59G06, Q99730 | Gene names | HTATSF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | HIV Tat-specific factor 1 (Tat-SF1). | |||||
|
TRDN_HUMAN
|
||||||
| NC score | 0.078707 (rank : 70) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13061 | Gene names | TRDN | |||
|
Domain Architecture |

|
|||||
| Description | Triadin. | |||||
|
SRCH_HUMAN
|
||||||
| NC score | 0.078111 (rank : 71) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P23327, Q504Y6 | Gene names | HRC, HCP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sarcoplasmic reticulum histidine-rich calcium-binding protein precursor. | |||||
|
SFRS4_HUMAN
|
||||||
| NC score | 0.078087 (rank : 72) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
NPBL_MOUSE
|
||||||
| NC score | 0.077715 (rank : 73) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q6KCD5, Q6KC78, Q7TNS4, Q8BKV4, Q8CES9, Q9CUC6 | Gene names | Nipbl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin homolog) (SCC2 homolog). | |||||
|
TARA_HUMAN
|
||||||
| NC score | 0.077651 (rank : 74) | θ value | 0.163984 (rank : 20) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q9H2D6, O94797, Q2PZW8, Q2Q3Z9, Q2Q400, Q5R3M6, Q96DW1, Q9BT77, Q9BTL7, Q9BY98, Q9Y3L4 | Gene names | TRIOBP, KIAA1662, TARA | |||
|
Domain Architecture |

|
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
CCD55_MOUSE
|
||||||
| NC score | 0.077550 (rank : 75) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 519 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q5NCR9 | Gene names | Ccdc55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 55. | |||||
|
ARS2_MOUSE
|
||||||
| NC score | 0.077304 (rank : 76) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99MR6, Q99MR4, Q99MR5, Q99MR7 | Gene names | Ars2, Asr2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
LRRF1_MOUSE
|
||||||
| NC score | 0.076958 (rank : 77) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 440 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q3UZ39, O70323, Q3TS94 | Gene names | Lrrfip1, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (FLI-LRR-associated protein 1) (Flap-1) (H186 FLAP). | |||||
|
SFR11_HUMAN
|
||||||
| NC score | 0.076492 (rank : 78) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q05519 | Gene names | SFRS11 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor arginine/serine-rich 11 (Arginine-rich 54 kDa nuclear protein) (p54). | |||||
|
SPRL1_MOUSE
|
||||||
| NC score | 0.076334 (rank : 79) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 403 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70663, P97810, Q99L82 | Gene names | Sparcl1, Ecm2, Sc1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (Matrix glycoprotein Sc1) (Extracellular matrix protein 2). | |||||
|
SIAL_HUMAN
|
||||||
| NC score | 0.076164 (rank : 80) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 77 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P21815 | Gene names | IBSP, BNSP | |||
|
Domain Architecture |

|
|||||
| Description | Bone sialoprotein-2 precursor (Bone sialoprotein II) (BSP II) (Cell- binding sialoprotein) (Integrin-binding sialoprotein). | |||||
|
NCKX1_HUMAN
|
||||||
| NC score | 0.076071 (rank : 81) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | O60721, O43485, O75184 | Gene names | SLC24A1, KIAA0702, NCKX1 | |||
|
Domain Architecture |

|
|||||
| Description | Sodium/potassium/calcium exchanger 1 (Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod Na-Ca+K exchanger). | |||||
|
CTR9_MOUSE
|
||||||
| NC score | 0.075776 (rank : 82) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q62018, Q3UFF5, Q3UY40, Q66JX4, Q7TPS6, Q8BND9, Q8BRD1, Q8C9W7, Q8C9Y3, Q8CHI1 | Gene names | Ctr9, Kiaa0155, Sh2bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1) (Tetratricopeptide repeat-containing, SH2-binding phosphoprotein of 150 kDa) (TPR-containing, SH2-binding phosphoprotein of 150 kDa) (p150TSP). | |||||
|
NIPBL_HUMAN
|
||||||
| NC score | 0.074814 (rank : 83) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1311 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6KC79, Q6KCD6, Q6N080, Q6ZT92, Q7Z2E6, Q8N4M5, Q9Y6Y3, Q9Y6Y4 | Gene names | NIPBL, IDN3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nipped-B-like protein (Delangin) (SCC2 homolog). | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.074527 (rank : 84) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
TARA_MOUSE
|
||||||
| NC score | 0.074383 (rank : 85) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1133 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99KW3, Q2PZW9, Q2Q402, Q2Q403, Q2Q404, Q6ZPK4, Q8C6T3, Q8CG90 | Gene names | Triobp, Kiaa1662, Tara | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TRIO and F-actin-binding protein (Protein Tara) (Trio-associated repeat on actin). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.073726 (rank : 86) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.073084 (rank : 87) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.072620 (rank : 88) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
ENL_HUMAN
|
||||||
| NC score | 0.071833 (rank : 89) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 305 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q03111, Q14768 | Gene names | MLLT1, ENL, LTG19, YEATS1 | |||
|
Domain Architecture |

|
|||||
| Description | Protein ENL (YEATS domain-containing protein 1). | |||||
|
RBBP6_MOUSE
|
||||||
| NC score | 0.071806 (rank : 90) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1014 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P97868, P70287, Q3TTR9, Q3TUM7, Q3UMP7, Q4U217, Q7TT06, Q8BNY8, Q8R399 | Gene names | Rbbp6, P2pr, Pact | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R). | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.071714 (rank : 91) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
CTR9_HUMAN
|
||||||
| NC score | 0.071297 (rank : 92) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q6PD62, Q15015 | Gene names | CTR9, KIAA0155, SH2BP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RNA polymerase-associated protein CTR9 homolog (SH2 domain-binding protein 1). | |||||
|
VCXC_HUMAN
|
||||||
| NC score | 0.070980 (rank : 93) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
AP3B1_HUMAN
|
||||||
| NC score | 0.070822 (rank : 94) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | O00203, O00580, Q7Z393, Q9HD66 | Gene names | AP3B1, ADTB3A | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
PRB4S_HUMAN
|
||||||
| NC score | 0.070488 (rank : 95) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P10163, P02813 | Gene names | PRB4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 4 allele S precursor (Salivary proline-rich protein Po) (Parotid o protein) [Contains: Protein N1; Glycosylated protein A]. | |||||
|
JARD2_MOUSE
|
||||||
| NC score | 0.070184 (rank : 96) | θ value | 0.62314 (rank : 26) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q62315, Q99LD1 | Gene names | Jarid2, Jmj | |||
|
Domain Architecture |

|
|||||
| Description | Jumonji protein (Jumonji/ARID domain-containing protein 2). | |||||
|
TCOF_MOUSE
|
||||||
| NC score | 0.070176 (rank : 97) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 471 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O08784, O08857 | Gene names | Tcof1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein homolog). | |||||
|
CMGA_HUMAN
|
||||||
| NC score | 0.069540 (rank : 98) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P10645, Q96E84, Q96GL7, Q9BQB5 | Gene names | CHGA | |||
|
Domain Architecture |

|
|||||
| Description | Chromogranin A precursor (CgA) (Pituitary secretory protein I) (SP-I) [Contains: Vasostatin-1 (Vasostatin I); Vasostatin-2 (Vasostatin II); EA-92; ES-43; Pancreastatin; SS-18; WA-8; WE-14; LF-19; AL-11; GV-19; GR-44; ER-37]. | |||||
|
ARS2_HUMAN
|
||||||
| NC score | 0.067668 (rank : 99) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9BXP5, O95808, Q9BWP6, Q9BXP4 | Gene names | ARS2, ASR2 | |||
|
Domain Architecture |

|
|||||
| Description | Arsenite-resistance protein 2. | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.067006 (rank : 100) | θ value | 0.00509761 (rank : 14) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
TRI44_HUMAN
|
||||||
| NC score | 0.066242 (rank : 101) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q96DX7, Q96QY2, Q9UGK0 | Gene names | TRIM44, DIPB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 44 (Protein DIPB). | |||||
|
SEMG2_HUMAN
|
||||||
| NC score | 0.065925 (rank : 102) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q02383, Q6X2M6 | Gene names | SEMG2 | |||
|
Domain Architecture |

|
|||||
| Description | Semenogelin-2 precursor (Semenogelin II) (SGII). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.065920 (rank : 103) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
VCX3_HUMAN
|
||||||
| NC score | 0.065598 (rank : 104) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
ZAR1_MOUSE
|
||||||
| NC score | 0.065188 (rank : 105) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 254 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q80SU3 | Gene names | Zar1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
SRRM2_HUMAN
|
||||||
| NC score | 0.063817 (rank : 106) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1296 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9UQ35, O15038, O94803, Q6NSL3, Q6PIM3, Q6PK40, Q8IW17, Q96GY7, Q9P0G1, Q9UHA8, Q9UQ36, Q9UQ37, Q9UQ38, Q9UQ40 | Gene names | SRRM2, KIAA0324, SRL300, SRM300 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2 (Serine/arginine-rich splicing factor-related nuclear matrix protein of 300 kDa) (Ser/Arg- related nuclear matrix protein) (SR-related nuclear matrix protein of 300 kDa) (Splicing coactivator subunit SRm300) (300 kDa nuclear matrix antigen). | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.063481 (rank : 107) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
CIR_HUMAN
|
||||||
| NC score | 0.063413 (rank : 108) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 249 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q86X95, O95367, Q12804, Q4G1B9, Q6PJI4, Q8IWI2 | Gene names | CIR | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CBF1-interacting corepressor (Recepin). | |||||
|
ZAR1_HUMAN
|
||||||
| NC score | 0.063055 (rank : 109) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q86SH2 | Gene names | ZAR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zygote arrest 1 (Oocyte-specific maternal effect factor). | |||||
|
DAXX_MOUSE
|
||||||
| NC score | 0.062056 (rank : 110) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O35613, Q9QWT8, Q9QWV3 | Gene names | Daxx | |||
|
Domain Architecture |

|
|||||
| Description | Death domain-associated protein 6 (Daxx). | |||||
|
TCAL5_HUMAN
|
||||||
| NC score | 0.061669 (rank : 111) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 806 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q5H9L2 | Gene names | TCEAL5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
TCAL3_MOUSE
|
||||||
| NC score | 0.061621 (rank : 112) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8R0A5, Q9CYK5, Q9D1S4 | Gene names | Tceal3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 3 (TCEA-like protein 3) (Transcription elongation factor S-II protein-like 3). | |||||
|
NNP1_MOUSE
|
||||||
| NC score | 0.061349 (rank : 113) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 47 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P56183, O35712, Q9ERE1, Q9JI07, Q9JK67, Q9JKU2 | Gene names | Nnp1 | |||
|
Domain Architecture |

|
|||||
| Description | NNP-1 protein (Novel nuclear protein 1) (Nop52). | |||||
|
CENPB_MOUSE
|
||||||
| NC score | 0.060255 (rank : 114) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P27790 | Gene names | Cenpb, Cenp-b | |||
|
Domain Architecture |

|
|||||
| Description | Major centromere autoantigen B (Centromere protein B) (CENP-B). | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.059522 (rank : 115) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.059368 (rank : 116) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
FUS_MOUSE
|
||||||
| NC score | 0.059191 (rank : 117) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 154 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P56959 | Gene names | Fus | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Pigpen protein). | |||||
|
MINT_MOUSE
|
||||||
| NC score | 0.058766 (rank : 118) | θ value | 5.27518 (rank : 55) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1514 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q62504, Q80TN9, Q99PS4, Q9QZW2 | Gene names | Spen, Kiaa0929, Mint, Sharp | |||
|
Domain Architecture |

|
|||||
| Description | Msx2-interacting protein (SPEN homolog) (SMART/HDAC1-associated repressor protein). | |||||
|
FUS_HUMAN
|
||||||
| NC score | 0.058298 (rank : 119) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P35637 | Gene names | FUS, TLS | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein FUS (Oncogene FUS) (Oncogene TLS) (Translocated in liposarcoma protein) (POMp75) (75 kDa DNA-pairing protein). | |||||
|
PPRB_MOUSE
|
||||||
| NC score | 0.058193 (rank : 120) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q925J9, O88323, Q3UHV0, Q6AXD5, Q8BW37, Q8BX19, Q8VDQ7, Q925K0 | Gene names | Pparbp, Trip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2). | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.058187 (rank : 121) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
TNC6B_HUMAN
|
||||||
| NC score | 0.057697 (rank : 122) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 256 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9UPQ9, Q5TH52, Q8TBX2 | Gene names | TNRC6B, KIAA1093 | |||
|
Domain Architecture |

|
|||||
| Description | Trinucleotide repeat-containing 6B protein. | |||||
|
PTMS_HUMAN
|
||||||
| NC score | 0.057483 (rank : 123) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20962 | Gene names | PTMS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
NALP5_MOUSE
|
||||||
| NC score | 0.057437 (rank : 124) | θ value | 0.21417 (rank : 21) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9R1M5, Q9JLR2 | Gene names | Nalp5, Mater | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 5 (Maternal antigen that embryos require) (Mater protein) (Ooplasm-specific protein 1) (OP1). | |||||
|
AFF4_HUMAN
|
||||||
| NC score | 0.057364 (rank : 125) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 611 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9UHB7, Q498B2, Q59FB3, Q6P592, Q8TDR1, Q9P0E4 | Gene names | AFF4, AF5Q31, MCEF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4 (ALL1-fused gene from chromosome 5q31) (Major CDK9 elongation factor-associated protein). | |||||
|
AP3B2_HUMAN
|
||||||
| NC score | 0.056778 (rank : 126) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q13367, O14808 | Gene names | AP3B2 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-2 (Adapter-related protein complex 3 beta-2 subunit) (Beta3B-adaptin) (Adaptor protein complex AP-3 beta-2 subunit) (AP-3 complex beta-2 subunit) (Clathrin assembly protein complex 3 beta-2 large chain) (Neuron-specific vesicle coat protein beta-NAP). | |||||
|
ACINU_HUMAN
|
||||||
| NC score | 0.056736 (rank : 127) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UKV3, O75158, Q9UG91, Q9UKV1, Q9UKV2 | Gene names | ACIN1, ACINUS, KIAA0670 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptotic chromatin condensation inducer in the nucleus (Acinus). | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.056480 (rank : 128) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
THOC2_HUMAN
|
||||||
| NC score | 0.055573 (rank : 129) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 337 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q8NI27, Q9H8I6 | Gene names | THOC2, CXorf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | THO complex subunit 2 (Tho2). | |||||
|
MAP1B_HUMAN
|
||||||
| NC score | 0.055359 (rank : 130) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | P46821 | Gene names | MAP1B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) [Contains: MAP1 light chain LC1]. | |||||
|
ATRX_MOUSE
|
||||||
| NC score | 0.055043 (rank : 131) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q61687 | Gene names | Atrx, Hp1bp2, Xnp | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked nuclear protein) (Heterochromatin protein 2) (HP1 alpha-interacting protein) (HP1-BP38 protein). | |||||
|
ATRX_HUMAN
|
||||||
| NC score | 0.054986 (rank : 132) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 842 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P46100, P51068, Q15886, Q59FB5, Q59H31, Q5H9A2, Q5JWI4, Q7Z2J1, Q9H0Z1, Q9NTS3 | Gene names | ATRX, RAD54L, XH2 | |||
|
Domain Architecture |

|
|||||
| Description | Transcriptional regulator ATRX (EC 3.6.1.-) (ATP-dependent helicase ATRX) (X-linked helicase II) (X-linked nuclear protein) (XNP) (Znf- HX). | |||||
|
GNAS3_MOUSE
|
||||||
| NC score | 0.054701 (rank : 133) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 84 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Z0F1, Q9QXW5, Q9Z0H2 | Gene names | Gnas, Gnas1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuroendocrine secretory protein 55 (NESP55) [Contains: LHAL tetrapeptide; GPIPIRRH peptide]. | |||||
|
TCAL5_MOUSE
|
||||||
| NC score | 0.054585 (rank : 134) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 63 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8CCT4, Q497R6 | Gene names | Tceal5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 5 (TCEA-like protein 5) (Transcription elongation factor S-II protein-like 5). | |||||
|
PRP5_HUMAN
|
||||||
| NC score | 0.054508 (rank : 135) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P04281 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide IB-1. | |||||
|
H13_MOUSE
|
||||||
| NC score | 0.053492 (rank : 136) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43277, Q8C6M4 | Gene names | Hist1h1d, H1f3 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.3 (H1 VAR.4) (H1d). | |||||
|
PRPE_HUMAN
|
||||||
| NC score | 0.053141 (rank : 137) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02811 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic proline-rich peptide P-E (IB-9). | |||||
|
NEUM_MOUSE
|
||||||
| NC score | 0.053060 (rank : 138) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P06837 | Gene names | Gap43, Basp2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuromodulin (Axonal membrane protein GAP-43) (Growth-associated protein 43) (Calmodulin-binding protein P-57). | |||||
|
AN32B_MOUSE
|
||||||
| NC score | 0.052734 (rank : 139) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9EST5 | Gene names | Anp32b, Pal31 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acidic leucine-rich nuclear phosphoprotein 32 family member B (Proliferation-related acidic leucine-rich protein PAL31). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.052620 (rank : 140) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
TXND2_MOUSE
|
||||||
| NC score | 0.052571 (rank : 141) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q6P902, Q8CJD1 | Gene names | Txndc2, Sptrx, Sptrx1, Trx4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thioredoxin domain-containing protein 2 (Spermatid-specific thioredoxin-1) (Sptrx-1) (Thioredoxin-4). | |||||
|
SRRM2_MOUSE
|
||||||
| NC score | 0.052536 (rank : 142) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BTI8, Q3TBY5, Q569P9, Q80U37, Q8K383 | Gene names | Srrm2, Kiaa0324 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 2. | |||||
|
PTMS_MOUSE
|
||||||
| NC score | 0.052039 (rank : 143) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9D0J8 | Gene names | Ptms | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Parathymosin. | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.051118 (rank : 144) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.051104 (rank : 145) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SH3BG_MOUSE
|
||||||
| NC score | 0.051001 (rank : 146) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 99 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9WUZ7 | Gene names | Sh3bgr | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding glutamic acid-rich protein (SH3BGR protein). | |||||
|
TCAL2_HUMAN
|
||||||
| NC score | 0.050919 (rank : 147) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 45 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9H3H9 | Gene names | TCEAL2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation factor A protein-like 2 (TCEA-like protein 2) (Transcription elongation factor S-II protein-like 2). | |||||
|
PPIG_HUMAN
|
||||||
| NC score | 0.050754 (rank : 148) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 670 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q13427, O00706, Q96DG9 | Gene names | PPIG | |||
|
Domain Architecture |

|
|||||
| Description | Peptidyl-prolyl cis-trans isomerase G (EC 5.2.1.8) (Peptidyl-prolyl isomerase G) (PPIase G) (Rotamase G) (Cyclophilin G) (Clk-associating RS-cyclophilin) (CARS-cyclophilin) (CARS-Cyp) (SR-cyclophilin) (SRcyp) (SR-cyp) (CASP10). | |||||
|
ZCH10_MOUSE
|
||||||
| NC score | 0.050203 (rank : 149) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9CX48 | Gene names | Zcchc10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 10. | |||||
|
H14_HUMAN
|
||||||
| NC score | 0.050021 (rank : 150) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P10412 | Gene names | HIST1H1E, H1F4 | |||
|
Domain Architecture |

|
|||||
| Description | Histone H1.4 (Histone H1b). | |||||
|
TR150_MOUSE
|
||||||
| NC score | 0.050020 (rank : 151) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 468 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q569Z6 | Gene names | Thrap3, Trap150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid hormone receptor-associated protein 3 (Thyroid hormone receptor-associated protein complex 150 kDa component) (Trap150). | |||||
|
DIDO1_HUMAN
|
||||||
| NC score | 0.048075 (rank : 152) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 960 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q9BTC0, O15043, Q3ZTL7, Q3ZTL8, Q4VXS1, Q4VXS2, Q96D72, Q9BQW0, Q9BW03, Q9H4G6, Q9H4G7, Q9NTU8, Q9NUM8, Q9UFB6 | Gene names | DIDO1, C20orf158, DATF1, KIAA0333 | |||
|
Domain Architecture |

|
|||||
| Description | Death-inducer obliterator 1 (DIO-1) (Death-associated transcription factor 1) (DATF-1) (hDido1). | |||||
|
MYST4_HUMAN
|
||||||
| NC score | 0.046624 (rank : 153) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 671 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q8WYB5, O15087, Q86Y05, Q8WU81, Q9UKW2, Q9UKW3, Q9UKX0 | Gene names | MYST4, KIAA0383, MORF, MOZ2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase MYST4 (EC 2.3.1.48) (EC 2.3.1.-) (MYST protein 4) (MOZ, YBF2/SAS3, SAS2 and TIP60 protein 4) (Histone acetyltransferase MOZ2) (Monocytic leukemia zinc finger protein- related factor) (Histone acetyltransferase MORF). | |||||
|
CJ086_HUMAN
|
||||||
| NC score | 0.045649 (rank : 154) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NXX6, Q5SQQ5, Q6P673, Q8WY66, Q9BS90 | Gene names | C10orf86 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf86. | |||||
|
RBM25_HUMAN
|
||||||
| NC score | 0.041805 (rank : 155) | θ value | 5.27518 (rank : 57) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 682 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P49756, Q9UEQ5, Q9UIE9 | Gene names | RBM25, RNPC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable RNA-binding protein 25 (RNA-binding motif protein 25) (RNA- binding region-containing protein 7) (Protein S164). | |||||
|
BTBDC_HUMAN
|
||||||
| NC score | 0.041284 (rank : 156) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8IY92, Q96JP1 | Gene names | BTBD12, KIAA1784 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | BTB/POZ domain-containing protein 12. | |||||
|
BRD4_MOUSE
|
||||||
| NC score | 0.037573 (rank : 157) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 845 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9ESU6, Q8VHF7, Q8VHF8 | Gene names | Brd4, Mcap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain-containing protein 4 (Mitotic chromosome-associated protein) (MCAP). | |||||
|
PESC_HUMAN
|
||||||
| NC score | 0.031912 (rank : 158) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00541, Q6IC29 | Gene names | PES1 | |||
|
Domain Architecture |

|
|||||
| Description | Pescadillo homolog 1. | |||||
|
PEG3_MOUSE
|
||||||
| NC score | 0.031662 (rank : 159) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1101 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q3URU2, O54978, Q3TQ69, Q5EBP7, Q61138, Q6GQS0, Q80U47, Q8R5N0, Q9QX53 | Gene names | Peg3, Kiaa0287, Pw1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein (ASF-1). | |||||
|
CLCB_HUMAN
|
||||||
| NC score | 0.031388 (rank : 160) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P09497, Q6FHW1 | Gene names | CLTB | |||
|
Domain Architecture |

|
|||||
| Description | Clathrin light chain B (Lcb). | |||||
|
LRC45_HUMAN
|
||||||
| NC score | 0.029381 (rank : 161) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 933 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96CN5 | Gene names | LRRC45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat-containing protein 45. | |||||
|
SPTB1_MOUSE
|
||||||
| NC score | 0.024257 (rank : 162) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P15508 | Gene names | Sptb, Spnb-1, Spnb1, Sptb1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.023266 (rank : 163) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
SPTB1_HUMAN
|
||||||
| NC score | 0.022449 (rank : 164) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P11277, Q15510, Q15519 | Gene names | SPTB, SPTB1 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, erythrocyte (Beta-I spectrin). | |||||
|
B3A3_HUMAN
|
||||||
| NC score | 0.020947 (rank : 165) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 119 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P48751 | Gene names | SLC4A3, AE3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3) (Cardiac/brain band 3-like protein) (CAE3/BAE3). | |||||
|
B3A3_MOUSE
|
||||||
| NC score | 0.020332 (rank : 166) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P16283 | Gene names | Slc4a3, Ae3 | |||
|
Domain Architecture |

|
|||||
| Description | Anion exchange protein 3 (Neuronal band 3-like protein) (Solute carrier family 4 member 3). | |||||
|
SMC4_MOUSE
|
||||||
| NC score | 0.019949 (rank : 167) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
PARPT_HUMAN
|
||||||
| NC score | 0.018840 (rank : 168) | θ value | 5.27518 (rank : 56) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7Z3E1, Q68CY9, Q86VP4, Q9Y4P7 | Gene names | TIPARP, PARP7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TCDD-inducible poly [ADP-ribose] polymerase (EC 2.4.2.30) (Poly [ADP- ribose] polymerase 7) (PARP-7). | |||||
|
PEG3_HUMAN
|
||||||
| NC score | 0.018203 (rank : 169) | θ value | 0.62314 (rank : 27) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9GZU2, P78418, Q5H9P9, Q7Z7H7, Q8TF75, Q9GZY2 | Gene names | PEG3, KIAA0287 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paternally expressed gene 3 protein. | |||||
|
K22O_HUMAN
|
||||||
| NC score | 0.013867 (rank : 170) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q01546, Q7Z795 | Gene names | KRT76, KRT2B, KRT2P | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 2 oral (Cytokeratin-2P) (K2P) (CK 2P) (Keratin 76). | |||||
|
ZKSC1_HUMAN
|
||||||
| NC score | 0.011063 (rank : 171) | θ value | 0.365318 (rank : 24) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P17029, P52745, Q8TBW5, Q8TEK7 | Gene names | ZKSCAN1, KOX18, ZNF139, ZNF36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger with KRAB and SCAN domain-containing protein 1 (Zinc finger protein 36) (Zinc finger protein KOX18). | |||||
|
ZBT17_HUMAN
|
||||||
| NC score | 0.010792 (rank : 172) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 925 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q13105, Q15932, Q5JYB2, Q9NUC9 | Gene names | ZBTB17, MIZ1, ZNF151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Myc-interacting zinc finger protein) (Miz-1 protein). | |||||
|
FGD4_MOUSE
|
||||||
| NC score | 0.009863 (rank : 173) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
ZBT17_MOUSE
|
||||||
| NC score | 0.009599 (rank : 174) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q60821, Q60699 | Gene names | Zbtb17, Zfp100, Znf151 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger and BTB domain-containing protein 17 (Zinc finger protein 151) (Zinc finger protein 100) (Zfp-100) (Polyomavirus late initiator promoter-binding protein) (LP-1) (Zinc finger protein Z13). | |||||
|
DDX55_MOUSE
|
||||||
| NC score | 0.009146 (rank : 175) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q6ZPL9, Q149H5, Q3U460, Q8BZR1 | Gene names | Ddx55, Kiaa1595 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-dependent RNA helicase DDX55 (EC 3.6.1.-) (DEAD box protein 55). | |||||
|
CAMKV_HUMAN
|
||||||
| NC score | 0.008889 (rank : 176) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1084 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q8NCB2, Q6FIB8, Q8NBS8, Q8NC85, Q8NDU4, Q8WTT8, Q9BQC9, Q9H0Q5 | Gene names | CAMKV | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CaM kinase-like vesicle-associated protein. | |||||
|
PTN18_MOUSE
|
||||||
| NC score | 0.005666 (rank : 177) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q61152, Q4JFH4, Q62404, Q922E3 | Gene names | Ptpn18, Flp1, Ptpk1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 18 (EC 3.1.3.48) (Fetal liver phosphatase 1) (FLP-1) (PTP-K1). | |||||
|
ABL1_HUMAN
|
||||||
| NC score | 0.004586 (rank : 178) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 1220 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P00519, Q13869, Q13870, Q16133, Q45F09 | Gene names | ABL1, ABL, JTK7 | |||
|
Domain Architecture |

|
|||||
| Description | Proto-oncogene tyrosine-protein kinase ABL1 (EC 2.7.10.2) (p150) (c- ABL) (Abelson murine leukemia viral oncogene homolog 1). | |||||
|
CORIN_MOUSE
|
||||||
| NC score | 0.002457 (rank : 179) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 70 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||