Please be patient as the page loads
|
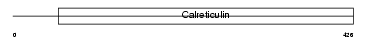
CALR3_HUMAN
|
||||||
| SwissProt Accessions | Q96L12, Q96LN3 | Gene names | CALR3, CRT2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
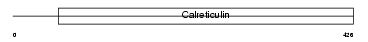
CALR3_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96L12, Q96LN3 | Gene names | CALR3, CRT2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
CALR3_MOUSE
|
||||||
| θ value | 3.82926e-184 (rank : 2) | NC score | 0.995576 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D9Q6 | Gene names | Calr3, Crt2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
CALR_HUMAN
|
||||||
| θ value | 2.45477e-106 (rank : 3) | NC score | 0.958911 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27797 | Gene names | CALR, CRTC | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) (grp60). | |||||
|
CALR_MOUSE
|
||||||
| θ value | 7.14227e-106 (rank : 4) | NC score | 0.962502 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14211 | Gene names | Calr | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60). | |||||
|
CALX_MOUSE
|
||||||
| θ value | 2.67802e-36 (rank : 5) | NC score | 0.829469 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35564 | Gene names | Canx | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor. | |||||
|
CLGN_HUMAN
|
||||||
| θ value | 1.32909e-35 (rank : 6) | NC score | 0.824015 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14967 | Gene names | CLGN | |||
|
Domain Architecture |
|
|||||
| Description | Calmegin precursor. | |||||
|
CALX_HUMAN
|
||||||
| θ value | 1.9192e-34 (rank : 7) | NC score | 0.822332 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
CLGN_MOUSE
|
||||||
| θ value | 3.06405e-32 (rank : 8) | NC score | 0.815561 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52194, Q9D2K5 | Gene names | Clgn, Meg1 | |||
|
Domain Architecture |
|
|||||
| Description | Calmegin precursor (MEG 1 antigen) (Calnexin-T) (A2/6). | |||||
|
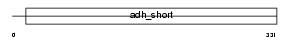
TGDS_MOUSE
|
||||||
| θ value | 0.125558 (rank : 9) | NC score | 0.039910 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VDR7 | Gene names | Tgds | |||
|
Domain Architecture |
|
|||||
| Description | dTDP-D-glucose 4,6-dehydratase (EC 4.2.1.46). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 0.47712 (rank : 10) | NC score | 0.012773 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.013637 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
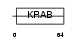
NR1D2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.006938 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14995, O00402, Q86XD4 | Gene names | NR1D2 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D2 (Rev-erb-beta) (EAR-1R) (Orphan nuclear hormone receptor BD73). | |||||
|
ZCHC7_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.026892 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N3Z6, Q5T0Q8, Q5T0Q9, Q5T0R0, Q8N2M1, Q8N4J2, Q8TBK8, Q9H648, Q9P0F0 | Gene names | ZCCHC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 7. | |||||
|
EVI1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 14) | NC score | 0.000552 (rank : 18) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||
|
PIBF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 15) | NC score | 0.006354 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
ZN155_HUMAN
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.000744 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12901, Q9UIE1, Q9UK14 | Gene names | ZNF155 | |||
|
Domain Architecture |
|
|||||
| Description | Zinc finger protein 155. | |||||
|
DEN2A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.005764 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULE3, Q1RMD5, Q86XY0 | Gene names | DENND2A, KIAA1277 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
ERIC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 18) | NC score | 0.077715 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
CALR3_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q96L12, Q96LN3 | Gene names | CALR3, CRT2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
CALR3_MOUSE
|
||||||
| NC score | 0.995576 (rank : 2) | θ value | 3.82926e-184 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9D9Q6 | Gene names | Calr3, Crt2 | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin-3 precursor (Calreticulin-2). | |||||
|
CALR_MOUSE
|
||||||
| NC score | 0.962502 (rank : 3) | θ value | 7.14227e-106 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 68 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P14211 | Gene names | Calr | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60). | |||||
|
CALR_HUMAN
|
||||||
| NC score | 0.958911 (rank : 4) | θ value | 2.45477e-106 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27797 | Gene names | CALR, CRTC | |||
|
Domain Architecture |

|
|||||
| Description | Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) (grp60). | |||||
|
CALX_MOUSE
|
||||||
| NC score | 0.829469 (rank : 5) | θ value | 2.67802e-36 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 76 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P35564 | Gene names | Canx | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor. | |||||
|
CLGN_HUMAN
|
||||||
| NC score | 0.824015 (rank : 6) | θ value | 1.32909e-35 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 111 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O14967 | Gene names | CLGN | |||
|
Domain Architecture |

|
|||||
| Description | Calmegin precursor. | |||||
|
CALX_HUMAN
|
||||||
| NC score | 0.822332 (rank : 7) | θ value | 1.9192e-34 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 96 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27824 | Gene names | CANX | |||
|
Domain Architecture |

|
|||||
| Description | Calnexin precursor (Major histocompatibility complex class I antigen- binding protein p88) (p90) (IP90). | |||||
|
CLGN_MOUSE
|
||||||
| NC score | 0.815561 (rank : 8) | θ value | 3.06405e-32 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 142 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P52194, Q9D2K5 | Gene names | Clgn, Meg1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmegin precursor (MEG 1 antigen) (Calnexin-T) (A2/6). | |||||
|
ERIC1_HUMAN
|
||||||
| NC score | 0.077715 (rank : 9) | θ value | θ > 10 (rank : 18) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q86X53, Q9P063 | Gene names | ERICH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glutamate-rich protein 1. | |||||
|
TGDS_MOUSE
|
||||||
| NC score | 0.039910 (rank : 10) | θ value | 0.125558 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VDR7 | Gene names | Tgds | |||
|
Domain Architecture |

|
|||||
| Description | dTDP-D-glucose 4,6-dehydratase (EC 4.2.1.46). | |||||
|
ZCHC7_HUMAN
|
||||||
| NC score | 0.026892 (rank : 11) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 73 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8N3Z6, Q5T0Q8, Q5T0Q9, Q5T0R0, Q8N2M1, Q8N4J2, Q8TBK8, Q9H648, Q9P0F0 | Gene names | ZCCHC7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCHC domain-containing protein 7. | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.013637 (rank : 12) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.012773 (rank : 13) | θ value | 0.47712 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
NR1D2_HUMAN
|
||||||
| NC score | 0.006938 (rank : 14) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14995, O00402, Q86XD4 | Gene names | NR1D2 | |||
|
Domain Architecture |

|
|||||
| Description | Orphan nuclear receptor NR1D2 (Rev-erb-beta) (EAR-1R) (Orphan nuclear hormone receptor BD73). | |||||
|
PIBF1_HUMAN
|
||||||
| NC score | 0.006354 (rank : 15) | θ value | 4.03905 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
DEN2A_HUMAN
|
||||||
| NC score | 0.005764 (rank : 16) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULE3, Q1RMD5, Q86XY0 | Gene names | DENND2A, KIAA1277 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DENN domain-containing protein 2A. | |||||
|
ZN155_HUMAN
|
||||||
| NC score | 0.000744 (rank : 17) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 786 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q12901, Q9UIE1, Q9UK14 | Gene names | ZNF155 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 155. | |||||
|
EVI1_HUMAN
|
||||||
| NC score | 0.000552 (rank : 18) | θ value | 4.03905 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03112, Q16122, Q99917 | Gene names | EVI1 | |||
|
Domain Architecture |

|
|||||
| Description | Ecotropic virus integration 1 site protein. | |||||