Please be patient as the page loads
|
B2L13_HUMAN
|
||||||
| SwissProt Accessions | Q9BXK5, Q96B37, Q96IB7, Q9BY01, Q9HC05, Q9UFE0, Q9UKN3 | Gene names | BCL2L13, MIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-like 13 protein (Protein Mil1) (Bcl-rambo). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
B2L13_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BXK5, Q96B37, Q96IB7, Q9BY01, Q9HC05, Q9UFE0, Q9UKN3 | Gene names | BCL2L13, MIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-like 13 protein (Protein Mil1) (Bcl-rambo). | |||||
|
B2L13_MOUSE
|
||||||
| θ value | 1.62381e-150 (rank : 2) | NC score | 0.959244 (rank : 2) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P59017 | Gene names | Bcl2l13, Mil1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-like 13 protein (Protein Mil1) (Bcl-rambo). | |||||
|
B2L14_HUMAN
|
||||||
| θ value | 0.00175202 (rank : 3) | NC score | 0.263022 (rank : 3) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZR8, Q96QR5, Q9BZR7 | Gene names | BCL2L14, BCLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis facilitator Bcl-2-like 14 protein (Apoptosis regulator Bcl- G). | |||||
|
B2L14_MOUSE
|
||||||
| θ value | 0.00869519 (rank : 4) | NC score | 0.232088 (rank : 4) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CPT0, Q3TVA7, Q8R141, Q9D3W3 | Gene names | Bcl2l14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis facilitator Bcl-2-like protein 14. | |||||
|
CSPG3_MOUSE
|
||||||
| θ value | 0.21417 (rank : 5) | NC score | 0.022906 (rank : 35) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
NFH_MOUSE
|
||||||
| θ value | 0.279714 (rank : 6) | NC score | 0.037244 (rank : 23) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
BCLX_MOUSE
|
||||||
| θ value | 0.365318 (rank : 7) | NC score | 0.097439 (rank : 5) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64373, O35844, Q60657, Q60658, Q61338 | Gene names | Bcl2l1, Bcl2l, Bclx | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis regulator Bcl-X (Bcl-2-like 1 protein). | |||||
|
SH23A_HUMAN
|
||||||
| θ value | 0.365318 (rank : 8) | NC score | 0.043274 (rank : 18) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BRG2, Q9Y2X4 | Gene names | SH2D3A, NSP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3A (Novel SH2-containing protein 1). | |||||
|
BCLX_HUMAN
|
||||||
| θ value | 0.47712 (rank : 9) | NC score | 0.095510 (rank : 6) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q07817, Q5TE65, Q92976 | Gene names | BCL2L1, BCL2L, BCLX | |||
|
Domain Architecture |
|
|||||
| Description | Apoptosis regulator Bcl-X (Bcl-2-like 1 protein). | |||||
|
NEST_HUMAN
|
||||||
| θ value | 0.47712 (rank : 10) | NC score | 0.030599 (rank : 28) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
PO121_HUMAN
|
||||||
| θ value | 1.06291 (rank : 11) | NC score | 0.056118 (rank : 12) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
ALO17_HUMAN
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.049812 (rank : 16) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
BCLW_HUMAN
|
||||||
| θ value | 1.38821 (rank : 13) | NC score | 0.091415 (rank : 7) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92843, Q5U0H4 | Gene names | BCL2L2, BCLW, KIAA0271 | |||
|
Domain Architecture |
|
|||||
| Description | Apoptosis regulator Bcl-W (Bcl-2-like 2 protein). | |||||
|
BCLW_MOUSE
|
||||||
| θ value | 1.38821 (rank : 14) | NC score | 0.089937 (rank : 8) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70345, Q545Q4, Q6A093, Q8CFR2, Q8CGL4, Q9CYW5 | Gene names | Bcl2l2, Bclw, Kiaa0271 | |||
|
Domain Architecture |
|
|||||
| Description | Apoptosis regulator Bcl-W (Bcl-2-like 2 protein) (c98). | |||||
|
LUZP1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 15) | NC score | 0.038690 (rank : 21) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
PTPRZ_HUMAN
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.012765 (rank : 51) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
STCH_MOUSE
|
||||||
| θ value | 1.81305 (rank : 17) | NC score | 0.017915 (rank : 42) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BM72, Q3TII6, Q9D1X5 | Gene names | Stch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 18) | NC score | 0.016031 (rank : 47) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
VCX1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 19) | NC score | 0.058384 (rank : 11) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
ZN294_HUMAN
|
||||||
| θ value | 2.36792 (rank : 20) | NC score | 0.031190 (rank : 27) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94822, Q9H8M4, Q9NUY5, Q9P0E9 | Gene names | ZNF294, C21orf10, C21orf98, KIAA0714 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 294. | |||||
|
APXL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 21) | NC score | 0.031775 (rank : 26) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | 3.0926 (rank : 22) | NC score | 0.027879 (rank : 32) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
MP2K5_MOUSE
|
||||||
| θ value | 3.0926 (rank : 23) | NC score | 0.003063 (rank : 58) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVS7, Q8CFM3, Q8K360, Q9D222 | Gene names | Map2k5, Mek5, Mkk5, Prkmk5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity mitogen-activated protein kinase kinase 5 (EC 2.7.12.2) (MAP kinase kinase 5) (MAPKK 5) (MAPK/ERK kinase 5). | |||||
|
RTN4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 24) | NC score | 0.029442 (rank : 31) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
VCX3_HUMAN
|
||||||
| θ value | 3.0926 (rank : 25) | NC score | 0.059298 (rank : 10) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
VCXC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 26) | NC score | 0.052585 (rank : 13) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 27) | NC score | 0.016230 (rank : 45) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
HGS_HUMAN
|
||||||
| θ value | 4.03905 (rank : 28) | NC score | 0.018197 (rank : 41) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
JHD1A_MOUSE
|
||||||
| θ value | 4.03905 (rank : 29) | NC score | 0.015188 (rank : 48) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59997, Q3U1M5, Q3UR56, Q3V3Q1, Q69ZT4 | Gene names | Fbxl11, Jhdm1a, Kiaa1004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1A (EC 1.14.11.-) (F-box/LRR-repeat protein 11) (F-box and leucine-rich repeat protein 11). | |||||
|
LAD1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 30) | NC score | 0.035839 (rank : 25) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
SNX17_HUMAN
|
||||||
| θ value | 4.03905 (rank : 31) | NC score | 0.040068 (rank : 19) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15036, Q53HN7, Q6IAS3 | Gene names | SNX17, KIAA0064 | |||
|
Domain Architecture |
|
|||||
| Description | Sorting nexin-17. | |||||
|
SNX17_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.039930 (rank : 20) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVL3, Q8R0N8 | Gene names | Snx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-17. | |||||
|
TMF1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.017635 (rank : 43) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
3BP5_MOUSE
|
||||||
| θ value | 5.27518 (rank : 34) | NC score | 0.037872 (rank : 22) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z131, Q8C903, Q8VCZ0 | Gene names | Sh3bp5, Sab | |||
|
Domain Architecture |
|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
CP007_MOUSE
|
||||||
| θ value | 5.27518 (rank : 35) | NC score | 0.024540 (rank : 33) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C190, Q6PFD1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 homolog (5-day ovary-specific transcript 1 protein). | |||||
|
FGD6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.016178 (rank : 46) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
PO121_MOUSE
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.045248 (rank : 17) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
PSMD6_MOUSE
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.023844 (rank : 34) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99JI4, Q9CWZ1 | Gene names | Psmd6 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 6 (26S proteasome regulatory subunit S10) (p42A). | |||||
|
3BP5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 39) | NC score | 0.036724 (rank : 24) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60239 | Gene names | SH3BP5, SAB | |||
|
Domain Architecture |
|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
ANLN_HUMAN
|
||||||
| θ value | 6.88961 (rank : 40) | NC score | 0.020198 (rank : 39) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
BAK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 41) | NC score | 0.079275 (rank : 9) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08734 | Gene names | Bak1, Bak | |||
|
Domain Architecture |
|
|||||
| Description | Bcl-2 homologous antagonist/killer (Apoptosis regulator BAK). | |||||
|
DNPEP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 42) | NC score | 0.021082 (rank : 36) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULA0, Q9BW44, Q9NUV5 | Gene names | DNPEP | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl aminopeptidase (EC 3.4.11.21). | |||||
|
K1024_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.018956 (rank : 40) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3V7, Q69ZS9 | Gene names | Kiaa1024 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0258 protein KIAA1024 (Protein DD1). | |||||
|
KI21A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.011346 (rank : 55) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
MAP4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.020926 (rank : 37) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
TIM_HUMAN
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.020444 (rank : 38) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNS1, O94802, Q86VM1, Q8IWH3 | Gene names | TIMELESS, TIM, TIM1, TIMELESS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Timeless homolog (hTIM). | |||||
|
ZMY12_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.030011 (rank : 30) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0C1, Q5VUS6, Q8TC87, Q96M51 | Gene names | ZMYND12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 12. | |||||
|
COG2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.016461 (rank : 44) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14746 | Gene names | COG2, LDLC | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 2 (Low density lipoprotein receptor defect C-complementing protein). | |||||
|
FBX30_MOUSE
|
||||||
| θ value | 8.99809 (rank : 49) | NC score | 0.013663 (rank : 50) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BJL1, Q8CI21, Q9D9X5 | Gene names | Fbxo30 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 30. | |||||
|
FYCO1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 50) | NC score | 0.014814 (rank : 49) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
HAPIP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 51) | NC score | 0.012005 (rank : 52) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60229 | Gene names | HAPIP, DUO | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein-interacting protein (Protein Duo). | |||||
|
K2C8_HUMAN
|
||||||
| θ value | 8.99809 (rank : 52) | NC score | 0.011762 (rank : 53) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P05787, Q14099, Q14716, Q14717, Q53GJ0, Q6DHW5, Q6GMY0, Q96J60 | Gene names | KRT8, CYK8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8). | |||||
|
K2C8_MOUSE
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.011629 (rank : 54) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P11679, Q3KQK5, Q3TGI1, Q3TJE1, Q3TKY7, Q61463, Q61518, Q61519 | Gene names | Krt8, Krt2-8 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8) (Cytokeratin endo A). | |||||
|
KI21A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.010557 (rank : 57) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
LAD1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.030183 (rank : 29) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
NEK4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.000550 (rank : 59) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1J2, O35673, Q9R1J1 | Gene names | Nek4, Stk2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek4 (EC 2.7.11.1) (NimA-related protein kinase 4) (Serine/threonine-protein kinase 2). | |||||
|
SYNE1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.011069 (rank : 56) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
BCL2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.052141 (rank : 14) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10415, P10416, Q13842, Q16197 | Gene names | BCL2 | |||
|
Domain Architecture |
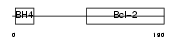
|
|||||
| Description | Apoptosis regulator Bcl-2. | |||||
|
BCL2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.051512 (rank : 15) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10417, P10418 | Gene names | Bcl2, Bcl-2 | |||
|
Domain Architecture |
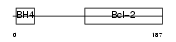
|
|||||
| Description | Apoptosis regulator Bcl-2. | |||||
|
B2L13_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9BXK5, Q96B37, Q96IB7, Q9BY01, Q9HC05, Q9UFE0, Q9UKN3 | Gene names | BCL2L13, MIL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-like 13 protein (Protein Mil1) (Bcl-rambo). | |||||
|
B2L13_MOUSE
|
||||||
| NC score | 0.959244 (rank : 2) | θ value | 1.62381e-150 (rank : 2) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P59017 | Gene names | Bcl2l13, Mil1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bcl-2-like 13 protein (Protein Mil1) (Bcl-rambo). | |||||
|
B2L14_HUMAN
|
||||||
| NC score | 0.263022 (rank : 3) | θ value | 0.00175202 (rank : 3) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9BZR8, Q96QR5, Q9BZR7 | Gene names | BCL2L14, BCLG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis facilitator Bcl-2-like 14 protein (Apoptosis regulator Bcl- G). | |||||
|
B2L14_MOUSE
|
||||||
| NC score | 0.232088 (rank : 4) | θ value | 0.00869519 (rank : 4) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CPT0, Q3TVA7, Q8R141, Q9D3W3 | Gene names | Bcl2l14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Apoptosis facilitator Bcl-2-like protein 14. | |||||
|
BCLX_MOUSE
|
||||||
| NC score | 0.097439 (rank : 5) | θ value | 0.365318 (rank : 7) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q64373, O35844, Q60657, Q60658, Q61338 | Gene names | Bcl2l1, Bcl2l, Bclx | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis regulator Bcl-X (Bcl-2-like 1 protein). | |||||
|
BCLX_HUMAN
|
||||||
| NC score | 0.095510 (rank : 6) | θ value | 0.47712 (rank : 9) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q07817, Q5TE65, Q92976 | Gene names | BCL2L1, BCL2L, BCLX | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis regulator Bcl-X (Bcl-2-like 1 protein). | |||||
|
BCLW_HUMAN
|
||||||
| NC score | 0.091415 (rank : 7) | θ value | 1.38821 (rank : 13) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q92843, Q5U0H4 | Gene names | BCL2L2, BCLW, KIAA0271 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis regulator Bcl-W (Bcl-2-like 2 protein). | |||||
|
BCLW_MOUSE
|
||||||
| NC score | 0.089937 (rank : 8) | θ value | 1.38821 (rank : 14) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 23 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P70345, Q545Q4, Q6A093, Q8CFR2, Q8CGL4, Q9CYW5 | Gene names | Bcl2l2, Bclw, Kiaa0271 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis regulator Bcl-W (Bcl-2-like 2 protein) (c98). | |||||
|
BAK_MOUSE
|
||||||
| NC score | 0.079275 (rank : 9) | θ value | 6.88961 (rank : 41) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O08734 | Gene names | Bak1, Bak | |||
|
Domain Architecture |

|
|||||
| Description | Bcl-2 homologous antagonist/killer (Apoptosis regulator BAK). | |||||
|
VCX3_HUMAN
|
||||||
| NC score | 0.059298 (rank : 10) | θ value | 3.0926 (rank : 25) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NNX9, Q9P0H4 | Gene names | VCX3A, VCX3, VCX8R, VCXA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 3 (VCX-A protein) (Variably charged protein X-A) (Variable charge protein on X with eight repeats) (VCX- 8r). | |||||
|
VCX1_HUMAN
|
||||||
| NC score | 0.058384 (rank : 11) | θ value | 2.36792 (rank : 19) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9H320, Q9P0H3 | Gene names | VCX, VCX1, VCX10R, VCXB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Variable charge X-linked protein 1 (VCX-B1 protein) (Variably charged protein X-B1) (Variable charge protein on X with ten repeats) (VCX- 10r). | |||||
|
PO121_HUMAN
|
||||||
| NC score | 0.056118 (rank : 12) | θ value | 1.06291 (rank : 11) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y2N3, O75115, Q9Y4S7 | Gene names | POM121, KIAA0618, NUP121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145). | |||||
|
VCXC_HUMAN
|
||||||
| NC score | 0.052585 (rank : 13) | θ value | 3.0926 (rank : 26) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9H321 | Gene names | VCXC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | VCX-C protein (Variably charged protein X-C). | |||||
|
BCL2_HUMAN
|
||||||
| NC score | 0.052141 (rank : 14) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 28 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10415, P10416, Q13842, Q16197 | Gene names | BCL2 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis regulator Bcl-2. | |||||
|
BCL2_MOUSE
|
||||||
| NC score | 0.051512 (rank : 15) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P10417, P10418 | Gene names | Bcl2, Bcl-2 | |||
|
Domain Architecture |

|
|||||
| Description | Apoptosis regulator Bcl-2. | |||||
|
ALO17_HUMAN
|
||||||
| NC score | 0.049812 (rank : 16) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 317 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9HCF4, Q69YK7, Q8IWF4, Q8IZX1, Q8IZX2, Q8N406 | Gene names | KIAA1618, ALO17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ALO17 (ALK lymphoma oligomerization partner on chromosome 17). | |||||
|
PO121_MOUSE
|
||||||
| NC score | 0.045248 (rank : 17) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8K3Z9, Q7TSH5 | Gene names | Pom121, Nup121 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa). | |||||
|
SH23A_HUMAN
|
||||||
| NC score | 0.043274 (rank : 18) | θ value | 0.365318 (rank : 8) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9BRG2, Q9Y2X4 | Gene names | SH2D3A, NSP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH2 domain-containing protein 3A (Novel SH2-containing protein 1). | |||||
|
SNX17_HUMAN
|
||||||
| NC score | 0.040068 (rank : 19) | θ value | 4.03905 (rank : 31) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15036, Q53HN7, Q6IAS3 | Gene names | SNX17, KIAA0064 | |||
|
Domain Architecture |

|
|||||
| Description | Sorting nexin-17. | |||||
|
SNX17_MOUSE
|
||||||
| NC score | 0.039930 (rank : 20) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BVL3, Q8R0N8 | Gene names | Snx17 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sorting nexin-17. | |||||
|
LUZP1_MOUSE
|
||||||
| NC score | 0.038690 (rank : 21) | θ value | 1.38821 (rank : 15) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 940 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8R4U7, Q3UV18, Q7TS71, Q8BQW1, Q99NG3, Q9CSL6 | Gene names | Luzp1, Luzp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine zipper protein 1 (Leucine zipper motif-containing protein). | |||||
|
3BP5_MOUSE
|
||||||
| NC score | 0.037872 (rank : 22) | θ value | 5.27518 (rank : 34) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Z131, Q8C903, Q8VCZ0 | Gene names | Sh3bp5, Sab | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
NFH_MOUSE
|
||||||
| NC score | 0.037244 (rank : 23) | θ value | 0.279714 (rank : 6) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 2247 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P19246, Q5SVF6, Q61959 | Gene names | Nefh, Nfh | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
3BP5_HUMAN
|
||||||
| NC score | 0.036724 (rank : 24) | θ value | 6.88961 (rank : 39) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 375 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O60239 | Gene names | SH3BP5, SAB | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
LAD1_HUMAN
|
||||||
| NC score | 0.035839 (rank : 25) | θ value | 4.03905 (rank : 30) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O00515, O95614 | Gene names | LAD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (120 kDa linear IgA bullous dermatosis antigen) (97 kDa linear IgA bullous dermatosis antigen) (Linear IgA disease antigen homolog) (LadA). | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.031775 (rank : 26) | θ value | 3.0926 (rank : 21) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
ZN294_HUMAN
|
||||||
| NC score | 0.031190 (rank : 27) | θ value | 2.36792 (rank : 20) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O94822, Q9H8M4, Q9NUY5, Q9P0E9 | Gene names | ZNF294, C21orf10, C21orf98, KIAA0714 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 294. | |||||
|
NEST_HUMAN
|
||||||
| NC score | 0.030599 (rank : 28) | θ value | 0.47712 (rank : 10) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1305 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P48681, O00552 | Gene names | NES | |||
|
Domain Architecture |

|
|||||
| Description | Nestin. | |||||
|
LAD1_MOUSE
|
||||||
| NC score | 0.030183 (rank : 29) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 391 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P57016 | Gene names | Lad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ladinin 1 (Lad-1) (Linear IgA disease autoantigen). | |||||
|
ZMY12_HUMAN
|
||||||
| NC score | 0.030011 (rank : 30) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H0C1, Q5VUS6, Q8TC87, Q96M51 | Gene names | ZMYND12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger MYND domain-containing protein 12. | |||||
|
RTN4_MOUSE
|
||||||
| NC score | 0.029442 (rank : 31) | θ value | 3.0926 (rank : 24) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q99P72, Q5DTK9, Q7TNB7, Q80W95, Q8BGK7, Q8BGM9, Q8K290, Q8K3G8, Q9CTE3 | Gene names | Rtn4, Kiaa0886, Nogo | |||
|
Domain Architecture |

|
|||||
| Description | Reticulon-4 (Neurite outgrowth inhibitor) (Nogo protein). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.027879 (rank : 32) | θ value | 3.0926 (rank : 22) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
CP007_MOUSE
|
||||||
| NC score | 0.024540 (rank : 33) | θ value | 5.27518 (rank : 35) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8C190, Q6PFD1 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C16orf7 homolog (5-day ovary-specific transcript 1 protein). | |||||
|
PSMD6_MOUSE
|
||||||
| NC score | 0.023844 (rank : 34) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99JI4, Q9CWZ1 | Gene names | Psmd6 | |||
|
Domain Architecture |

|
|||||
| Description | 26S proteasome non-ATPase regulatory subunit 6 (26S proteasome regulatory subunit S10) (p42A). | |||||
|
CSPG3_MOUSE
|
||||||
| NC score | 0.022906 (rank : 35) | θ value | 0.21417 (rank : 5) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 657 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P55066, Q6P1E3, Q8C4F8 | Gene names | Cspg3, Ncan | |||
|
Domain Architecture |

|
|||||
| Description | Neurocan core protein precursor (Chondroitin sulfate proteoglycan 3). | |||||
|
DNPEP_HUMAN
|
||||||
| NC score | 0.021082 (rank : 36) | θ value | 6.88961 (rank : 42) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9ULA0, Q9BW44, Q9NUV5 | Gene names | DNPEP | |||
|
Domain Architecture |

|
|||||
| Description | Aspartyl aminopeptidase (EC 3.4.11.21). | |||||
|
MAP4_HUMAN
|
||||||
| NC score | 0.020926 (rank : 37) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 428 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P27816, Q13082, Q96A76 | Gene names | MAP4 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 4 (MAP 4). | |||||
|
TIM_HUMAN
|
||||||
| NC score | 0.020444 (rank : 38) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UNS1, O94802, Q86VM1, Q8IWH3 | Gene names | TIMELESS, TIM, TIM1, TIMELESS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Timeless homolog (hTIM). | |||||
|
ANLN_HUMAN
|
||||||
| NC score | 0.020198 (rank : 39) | θ value | 6.88961 (rank : 40) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 222 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NQW6, Q5CZ78, Q6NSK5, Q9H8Y4, Q9NVN9, Q9NVP0 | Gene names | ANLN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Actin-binding protein anillin. | |||||
|
K1024_MOUSE
|
||||||
| NC score | 0.018956 (rank : 40) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K3V7, Q69ZS9 | Gene names | Kiaa1024 | |||
|
Domain Architecture |

|
|||||
| Description | UPF0258 protein KIAA1024 (Protein DD1). | |||||
|
HGS_HUMAN
|
||||||
| NC score | 0.018197 (rank : 41) | θ value | 4.03905 (rank : 28) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 517 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14964, Q9NR36 | Gene names | HGS, HRS | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hepatocyte growth factor-regulated tyrosine kinase substrate (Protein pp110) (Hrs). | |||||
|
STCH_MOUSE
|
||||||
| NC score | 0.017915 (rank : 42) | θ value | 1.81305 (rank : 17) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8BM72, Q3TII6, Q9D1X5 | Gene names | Stch | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Stress 70 protein chaperone microsome-associated 60 kDa protein precursor (Microsomal stress 70 protein ATPase core). | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.017635 (rank : 43) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
COG2_HUMAN
|
||||||
| NC score | 0.016461 (rank : 44) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 35 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q14746 | Gene names | COG2, LDLC | |||
|
Domain Architecture |

|
|||||
| Description | Conserved oligomeric Golgi complex component 2 (Low density lipoprotein receptor defect C-complementing protein). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.016230 (rank : 45) | θ value | 4.03905 (rank : 27) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
FGD6_MOUSE
|
||||||
| NC score | 0.016178 (rank : 46) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q69ZL1, Q8C8W5, Q8K3B0, Q9D3Y7 | Gene names | Fgd6, Kiaa1362 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 6. | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.016031 (rank : 47) | θ value | 2.36792 (rank : 18) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
JHD1A_MOUSE
|
||||||
| NC score | 0.015188 (rank : 48) | θ value | 4.03905 (rank : 29) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P59997, Q3U1M5, Q3UR56, Q3V3Q1, Q69ZT4 | Gene names | Fbxl11, Jhdm1a, Kiaa1004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | JmjC domain-containing histone demethylation protein 1A (EC 1.14.11.-) (F-box/LRR-repeat protein 11) (F-box and leucine-rich repeat protein 11). | |||||
|
FYCO1_MOUSE
|
||||||
| NC score | 0.014814 (rank : 49) | θ value | 8.99809 (rank : 50) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8VDC1, Q3V385, Q7TMT0, Q8BJN2 | Gene names | Fyco1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1. | |||||
|
FBX30_MOUSE
|
||||||
| NC score | 0.013663 (rank : 50) | θ value | 8.99809 (rank : 49) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BJL1, Q8CI21, Q9D9X5 | Gene names | Fbxo30 | |||
|
Domain Architecture |

|
|||||
| Description | F-box only protein 30. | |||||
|
PTPRZ_HUMAN
|
||||||
| NC score | 0.012765 (rank : 51) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 255 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P23471 | Gene names | PTPRZ1, PTPRZ, PTPZ | |||
|
Domain Architecture |

|
|||||
| Description | Receptor-type tyrosine-protein phosphatase zeta precursor (EC 3.1.3.48) (R-PTP-zeta). | |||||
|
HAPIP_HUMAN
|
||||||
| NC score | 0.012005 (rank : 52) | θ value | 8.99809 (rank : 51) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O60229 | Gene names | HAPIP, DUO | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-associated protein-interacting protein (Protein Duo). | |||||
|
K2C8_HUMAN
|
||||||
| NC score | 0.011762 (rank : 53) | θ value | 8.99809 (rank : 52) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P05787, Q14099, Q14716, Q14717, Q53GJ0, Q6DHW5, Q6GMY0, Q96J60 | Gene names | KRT8, CYK8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8). | |||||
|
K2C8_MOUSE
|
||||||
| NC score | 0.011629 (rank : 54) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P11679, Q3KQK5, Q3TGI1, Q3TJE1, Q3TKY7, Q61463, Q61518, Q61519 | Gene names | Krt8, Krt2-8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8) (Cytokeratin endo A). | |||||
|
KI21A_MOUSE
|
||||||
| NC score | 0.011346 (rank : 55) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9QXL2, Q6P5H1, Q6ZPJ8, Q8BWZ9, Q8BXF1 | Gene names | Kif21a, Kiaa1708 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A. | |||||
|
SYNE1_HUMAN
|
||||||
| NC score | 0.011069 (rank : 56) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1484 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8NF91, O94890, Q5JV19, Q5JV22, Q8N9P7, Q8TCP1, Q8WWW6, Q8WWW7, Q8WXF6, Q96N17, Q9C0A7, Q9H525, Q9H526, Q9NS36, Q9NU50, Q9UJ06, Q9UJ07, Q9ULF8 | Gene names | SYNE1, KIAA0796, KIAA1262, KIAA1756, MYNE1 | |||
|
Domain Architecture |

|
|||||
| Description | Nesprin-1 (Nuclear envelope spectrin repeat protein 1) (Synaptic nuclear envelope protein 1) (Syne-1) (Myocyte nuclear envelope protein 1) (Myne-1) (Enaptin). | |||||
|
KI21A_HUMAN
|
||||||
| NC score | 0.010557 (rank : 57) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 1543 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q7Z4S6, Q6UKL9, Q7Z668, Q86WZ5, Q8IVZ8, Q9C0F5, Q9NXU4, Q9Y590 | Gene names | KIF21A, KIAA1708, KIF2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin family member 21A (Kinesin-like protein KIF2) (NY-REN-62 antigen). | |||||
|
MP2K5_MOUSE
|
||||||
| NC score | 0.003063 (rank : 58) | θ value | 3.0926 (rank : 23) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 840 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WVS7, Q8CFM3, Q8K360, Q9D222 | Gene names | Map2k5, Mek5, Mkk5, Prkmk5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual specificity mitogen-activated protein kinase kinase 5 (EC 2.7.12.2) (MAP kinase kinase 5) (MAPKK 5) (MAPK/ERK kinase 5). | |||||
|
NEK4_MOUSE
|
||||||
| NC score | 0.000550 (rank : 59) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 57 | Target Neighborhood Hits | 899 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z1J2, O35673, Q9R1J1 | Gene names | Nek4, Stk2 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase Nek4 (EC 2.7.11.1) (NimA-related protein kinase 4) (Serine/threonine-protein kinase 2). | |||||