Please be patient as the page loads
|
VTI1B_HUMAN
|
||||||
| SwissProt Accessions | Q9UEU0, O43547 | Gene names | VTI1B, VTI1, VTI1L, VTI1L1, VTI2 | |||
|
Domain Architecture |
|
|||||
| Description | Vesicle transport through interaction with t-SNAREs homolog 1B (Vesicle transport v-SNARE protein Vti1-like 1) (Vti1-rp1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
VTI1B_HUMAN
|
||||||
| θ value | 4.94618e-107 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UEU0, O43547 | Gene names | VTI1B, VTI1, VTI1L, VTI1L1, VTI2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle transport through interaction with t-SNAREs homolog 1B (Vesicle transport v-SNARE protein Vti1-like 1) (Vti1-rp1). | |||||
|
VTI1B_MOUSE
|
||||||
| θ value | 8.4565e-99 (rank : 2) | NC score | 0.988224 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88384 | Gene names | Vti1b, Vti1l1 | |||
|
Domain Architecture |
|
|||||
| Description | Vesicle transport through interaction with t-SNAREs homolog 1B (Vesicle transport v-SNARE protein Vti1-like 1) (Vti1-rp1). | |||||
|
VTI1A_MOUSE
|
||||||
| θ value | 1.058e-16 (rank : 3) | NC score | 0.704777 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O89116, Q545P9 | Gene names | Vti1a, Vti1, Vti1l2 | |||
|
Domain Architecture |
|
|||||
| Description | Vesicle transport through interaction with t-SNAREs homolog 1A (Vesicle transport v-SNARE protein Vti1-like 2) (Vti1-rp2). | |||||
|
VTI1A_HUMAN
|
||||||
| θ value | 3.07829e-16 (rank : 4) | NC score | 0.697807 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96AJ9 | Gene names | VTI1A | |||
|
Domain Architecture |
|
|||||
| Description | Vesicle transport through interaction with t-SNAREs homolog 1A (Vesicle transport v-SNARE protein Vti1-like 2) (Vti1-rp2). | |||||
|
UACA_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 5) | NC score | 0.064930 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
CN045_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 6) | NC score | 0.138200 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8ND07, Q9H5P8 | Gene names | C14orf45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf45. | |||||
|
SLK_HUMAN
|
||||||
| θ value | 0.163984 (rank : 7) | NC score | 0.031447 (rank : 51) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
SLK_MOUSE
|
||||||
| θ value | 0.279714 (rank : 8) | NC score | 0.030225 (rank : 53) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
GOGA2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.073716 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 0.365318 (rank : 10) | NC score | 0.070256 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | 0.47712 (rank : 11) | NC score | 0.063021 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
SAPS1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 12) | NC score | 0.030127 (rank : 54) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
SNP29_HUMAN
|
||||||
| θ value | 0.62314 (rank : 13) | NC score | 0.112100 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95721 | Gene names | SNAP29 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptosomal-associated protein 29 (SNAP-29) (Vesicle-membrane fusion protein SNAP-29) (Soluble 29 kDa NSF attachment protein). | |||||
|
ROCK1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 14) | NC score | 0.030628 (rank : 52) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 15) | NC score | 0.065846 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
K2C8_MOUSE
|
||||||
| θ value | 1.38821 (rank : 16) | NC score | 0.032992 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P11679, Q3KQK5, Q3TGI1, Q3TJE1, Q3TKY7, Q61463, Q61518, Q61519 | Gene names | Krt8, Krt2-8 | |||
|
Domain Architecture |
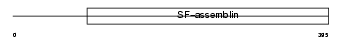
|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8) (Cytokeratin endo A). | |||||
|
LZTS2_HUMAN
|
||||||
| θ value | 1.38821 (rank : 17) | NC score | 0.065744 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BRK4, Q8N3I0, Q96J79, Q96JL2 | Gene names | LZTS2, KIAA1813, LAPSER1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2 (Protein LAPSER1). | |||||
|
A4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 18) | NC score | 0.043790 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
ANR35_HUMAN
|
||||||
| θ value | 1.81305 (rank : 19) | NC score | 0.035942 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 20) | NC score | 0.067738 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
SNP29_MOUSE
|
||||||
| θ value | 1.81305 (rank : 21) | NC score | 0.099612 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ERB0, Q3UGN0, Q9DBC5 | Gene names | Snap29 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptosomal-associated protein 29 (SNAP-29) (Vesicle-membrane fusion protein SNAP-29) (Soluble 29 kDa NSF attachment protein) (Golgi SNARE of 32 kDa) (Gs32). | |||||
|
BFSP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 22) | NC score | 0.036168 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13515, Q9HBW5 | Gene names | BFSP2 | |||
|
Domain Architecture |
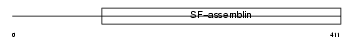
|
|||||
| Description | Phakinin (Beaded filament structural protein 2) (Lens fiber cell beaded filament protein CP 49) (CP49) (49 kDa cytoskeletal protein) (CP 47) (CP47) (Lens intermediate filament-like light) (LIFL-L). | |||||
|
CASP_HUMAN
|
||||||
| θ value | 2.36792 (rank : 23) | NC score | 0.058835 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 2.36792 (rank : 24) | NC score | 0.060303 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 25) | NC score | 0.053644 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
K2C8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 26) | NC score | 0.032979 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P05787, Q14099, Q14716, Q14717, Q53GJ0, Q6DHW5, Q6GMY0, Q96J60 | Gene names | KRT8, CYK8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8). | |||||
|
REST_MOUSE
|
||||||
| θ value | 2.36792 (rank : 27) | NC score | 0.053988 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
SNX23_HUMAN
|
||||||
| θ value | 2.36792 (rank : 28) | NC score | 0.037999 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 3.0926 (rank : 29) | NC score | 0.058700 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
MYH4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 30) | NC score | 0.040681 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 3.0926 (rank : 31) | NC score | 0.053329 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
SAS6_MOUSE
|
||||||
| θ value | 4.03905 (rank : 32) | NC score | 0.055103 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
SEC20_MOUSE
|
||||||
| θ value | 4.03905 (rank : 33) | NC score | 0.051339 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6QD59, Q5M9M2 | Gene names | Bnip1, Sec20, Sec20l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vesicle transport protein SEC20. | |||||
|
SNP23_HUMAN
|
||||||
| θ value | 4.03905 (rank : 34) | NC score | 0.055327 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00161, O00162, Q13602, Q6IAE3 | Gene names | SNAP23 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptosomal-associated protein 23 (SNAP-23) (Vesicle-membrane fusion protein SNAP-23). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 4.03905 (rank : 35) | NC score | 0.057472 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 36) | NC score | 0.059269 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
CP135_HUMAN
|
||||||
| θ value | 5.27518 (rank : 37) | NC score | 0.052453 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CTGE4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 38) | NC score | 0.034095 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IX94, O95046 | Gene names | CTAGE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 4 (cTAGE-4 protein). | |||||
|
CTTB2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 39) | NC score | 0.029834 (rank : 55) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | 5.27518 (rank : 40) | NC score | 0.055240 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
LAMA4_HUMAN
|
||||||
| θ value | 5.27518 (rank : 41) | NC score | 0.031955 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16363, Q14735, Q15335, Q4LE44, Q5SZG8, Q9UE18, Q9UJN9 | Gene names | LAMA4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
MYO5A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 42) | NC score | 0.032032 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
A4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 43) | NC score | 0.038659 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
CP135_MOUSE
|
||||||
| θ value | 6.88961 (rank : 44) | NC score | 0.053427 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.054067 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 8.99809 (rank : 46) | NC score | 0.046846 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
TRIM2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 47) | NC score | 0.012301 (rank : 57) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C040, O60272, Q9BSI9, Q9UFZ1 | Gene names | TRIM2, KIAA0517, RNF86 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 2 (RING finger protein 86). | |||||
|
TRIM2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 48) | NC score | 0.012340 (rank : 56) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESN6, Q3UHP5, Q8C981 | Gene names | Trim2, Narf | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 2 (Neural activity-related RING finger protein). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.053123 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
CT2NL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.050177 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.050358 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
GOG8A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.052761 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 635 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZW0, O94937, Q9NZG8, Q9NZW3 | Gene names | GOLGA8A, KIAA0855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 8A/B (Golgi autoantigen golgin-67) (88 kDa Golgi protein) (Gm88 autoantigen). | |||||
|
GOGA2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.058117 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
IFT74_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.054707 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.054300 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
K0555_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.050360 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
RAD50_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.063215 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
VTI1B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.94618e-107 (rank : 1) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 48 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9UEU0, O43547 | Gene names | VTI1B, VTI1, VTI1L, VTI1L1, VTI2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle transport through interaction with t-SNAREs homolog 1B (Vesicle transport v-SNARE protein Vti1-like 1) (Vti1-rp1). | |||||
|
VTI1B_MOUSE
|
||||||
| NC score | 0.988224 (rank : 2) | θ value | 8.4565e-99 (rank : 2) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 34 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | O88384 | Gene names | Vti1b, Vti1l1 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle transport through interaction with t-SNAREs homolog 1B (Vesicle transport v-SNARE protein Vti1-like 1) (Vti1-rp1). | |||||
|
VTI1A_MOUSE
|
||||||
| NC score | 0.704777 (rank : 3) | θ value | 1.058e-16 (rank : 3) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O89116, Q545P9 | Gene names | Vti1a, Vti1, Vti1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle transport through interaction with t-SNAREs homolog 1A (Vesicle transport v-SNARE protein Vti1-like 2) (Vti1-rp2). | |||||
|
VTI1A_HUMAN
|
||||||
| NC score | 0.697807 (rank : 4) | θ value | 3.07829e-16 (rank : 4) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 39 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96AJ9 | Gene names | VTI1A | |||
|
Domain Architecture |

|
|||||
| Description | Vesicle transport through interaction with t-SNAREs homolog 1A (Vesicle transport v-SNARE protein Vti1-like 2) (Vti1-rp2). | |||||
|
CN045_HUMAN
|
||||||
| NC score | 0.138200 (rank : 5) | θ value | 0.0961366 (rank : 6) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8ND07, Q9H5P8 | Gene names | C14orf45 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C14orf45. | |||||
|
SNP29_HUMAN
|
||||||
| NC score | 0.112100 (rank : 6) | θ value | 0.62314 (rank : 13) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O95721 | Gene names | SNAP29 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptosomal-associated protein 29 (SNAP-29) (Vesicle-membrane fusion protein SNAP-29) (Soluble 29 kDa NSF attachment protein). | |||||
|
SNP29_MOUSE
|
||||||
| NC score | 0.099612 (rank : 7) | θ value | 1.81305 (rank : 21) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9ERB0, Q3UGN0, Q9DBC5 | Gene names | Snap29 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptosomal-associated protein 29 (SNAP-29) (Vesicle-membrane fusion protein SNAP-29) (Soluble 29 kDa NSF attachment protein) (Golgi SNARE of 32 kDa) (Gs32). | |||||
|
GOGA2_HUMAN
|
||||||
| NC score | 0.073716 (rank : 8) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.070256 (rank : 9) | θ value | 0.365318 (rank : 10) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.067738 (rank : 10) | θ value | 1.81305 (rank : 20) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.065846 (rank : 11) | θ value | 1.06291 (rank : 15) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
LZTS2_HUMAN
|
||||||
| NC score | 0.065744 (rank : 12) | θ value | 1.38821 (rank : 17) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9BRK4, Q8N3I0, Q96J79, Q96JL2 | Gene names | LZTS2, KIAA1813, LAPSER1 | |||
|
Domain Architecture |

|
|||||
| Description | Leucine zipper putative tumor suppressor 2 (Protein LAPSER1). | |||||
|
UACA_MOUSE
|
||||||
| NC score | 0.064930 (rank : 13) | θ value | 0.0252991 (rank : 5) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q8CGB3, Q69ZG3, Q7TN77, Q8BJC8 | Gene names | Uaca, Kiaa1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein (Nucling) (Nuclear membrane-binding protein). | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.063215 (rank : 14) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.063021 (rank : 15) | θ value | 0.47712 (rank : 11) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.060303 (rank : 16) | θ value | 2.36792 (rank : 24) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.059269 (rank : 17) | θ value | 5.27518 (rank : 36) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
CASP_HUMAN
|
||||||
| NC score | 0.058835 (rank : 18) | θ value | 2.36792 (rank : 23) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q13948, Q53GU9, Q8TBS3 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.058700 (rank : 19) | θ value | 3.0926 (rank : 29) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
GOGA2_MOUSE
|
||||||
| NC score | 0.058117 (rank : 20) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.057472 (rank : 21) | θ value | 4.03905 (rank : 35) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
SNP23_HUMAN
|
||||||
| NC score | 0.055327 (rank : 22) | θ value | 4.03905 (rank : 34) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O00161, O00162, Q13602, Q6IAE3 | Gene names | SNAP23 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptosomal-associated protein 23 (SNAP-23) (Vesicle-membrane fusion protein SNAP-23). | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.055240 (rank : 23) | θ value | 5.27518 (rank : 40) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
SAS6_MOUSE
|
||||||
| NC score | 0.055103 (rank : 24) | θ value | 4.03905 (rank : 32) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.054707 (rank : 25) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.054300 (rank : 26) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.054067 (rank : 27) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.053988 (rank : 28) | θ value | 2.36792 (rank : 27) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.053644 (rank : 29) | θ value | 2.36792 (rank : 25) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.053427 (rank : 30) | θ value | 6.88961 (rank : 44) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.053329 (rank : 31) | θ value | 3.0926 (rank : 31) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.053123 (rank : 32) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
GOG8A_HUMAN
|
||||||
| NC score | 0.052761 (rank : 33) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 635 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9NZW0, O94937, Q9NZG8, Q9NZW3 | Gene names | GOLGA8A, KIAA0855 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 8A/B (Golgi autoantigen golgin-67) (88 kDa Golgi protein) (Gm88 autoantigen). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.052453 (rank : 34) | θ value | 5.27518 (rank : 37) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
SEC20_MOUSE
|
||||||
| NC score | 0.051339 (rank : 35) | θ value | 4.03905 (rank : 33) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6QD59, Q5M9M2 | Gene names | Bnip1, Sec20, Sec20l | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Vesicle transport protein SEC20. | |||||
|
K0555_HUMAN
|
||||||
| NC score | 0.050360 (rank : 36) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.050358 (rank : 37) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
CT2NL_HUMAN
|
||||||
| NC score | 0.050177 (rank : 38) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1059 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q9P2B4, Q96B40 | Gene names | CTTNBP2NL, KIAA1433 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CTTNBP2 N-terminal-like protein. | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.046846 (rank : 39) | θ value | 8.99809 (rank : 46) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
A4_HUMAN
|
||||||
| NC score | 0.043790 (rank : 40) | θ value | 1.81305 (rank : 18) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 274 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P05067, P09000, P78438, Q13764, Q13778, Q13793, Q16011, Q16014, Q16019, Q16020, Q9BT38, Q9UCA9, Q9UCB6, Q9UCC8, Q9UCD1, Q9UQ58 | Gene names | APP, A4, AD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein) (Cerebral vascular amyloid peptide) (CVAP) (Protease nexin-II) (PN-II) (APPI) (PreA4) [Contains: Soluble APP-alpha (S-APP- alpha); Soluble APP-beta (S-APP-beta); C99; Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)); Gamma-CTF(57) (Gamma- secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
MYH4_MOUSE
|
||||||
| NC score | 0.040681 (rank : 41) | θ value | 3.0926 (rank : 30) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1476 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q5SX39 | Gene names | Myh4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-4 (Myosin heavy chain, skeletal muscle, fetal). | |||||
|
A4_MOUSE
|
||||||
| NC score | 0.038659 (rank : 42) | θ value | 6.88961 (rank : 43) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P12023, P97487, P97942, Q99K32 | Gene names | App | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amyloid beta A4 protein precursor (APP) (ABPP) (Alzheimer disease amyloid protein homolog) (Amyloidogenic glycoprotein) (AG) [Contains: Soluble APP-alpha (S-APP-alpha); Soluble APP-beta (S-APP-beta); C99 (APP-C99); Beta-amyloid protein 42 (Beta-APP42); Beta-amyloid protein 40 (Beta-APP40); C83; P3(42); P3(40); Gamma-CTF(59) (Gamma-secretase C-terminal fragment 59) (Amyloid intracellular domain 59) (AID(59)) (APP-C59); Gamma-CTF(57) (Gamma-secretase C-terminal fragment 57) (Amyloid intracellular domain 57) (AID(57)) (APP-C57); Gamma-CTF(50) (Gamma-secretase C-terminal fragment 50) (Amyloid intracellular domain 50) (AID(50)); C31]. | |||||
|
SNX23_HUMAN
|
||||||
| NC score | 0.037999 (rank : 43) | θ value | 2.36792 (rank : 28) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q96L93, Q5HYC0, Q5HYK1, Q5JWW3, Q5TFK5, Q86VL9, Q86YS5, Q8IYU0, Q9BQJ8, Q9BQM0, Q9BQM1, Q9BQM5, Q9H5U0, Q9HCI2, Q9NXN9 | Gene names | C20orf23, KIAA1590, SNX23 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin-like motor protein C20orf23 (Sorting nexin-23). | |||||
|
BFSP2_HUMAN
|
||||||
| NC score | 0.036168 (rank : 44) | θ value | 2.36792 (rank : 22) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q13515, Q9HBW5 | Gene names | BFSP2 | |||
|
Domain Architecture |

|
|||||
| Description | Phakinin (Beaded filament structural protein 2) (Lens fiber cell beaded filament protein CP 49) (CP49) (49 kDa cytoskeletal protein) (CP 47) (CP47) (Lens intermediate filament-like light) (LIFL-L). | |||||
|
ANR35_HUMAN
|
||||||
| NC score | 0.035942 (rank : 45) | θ value | 1.81305 (rank : 19) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
CTGE4_HUMAN
|
||||||
| NC score | 0.034095 (rank : 46) | θ value | 5.27518 (rank : 38) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 393 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q8IX94, O95046 | Gene names | CTAGE4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 4 (cTAGE-4 protein). | |||||
|
K2C8_MOUSE
|
||||||
| NC score | 0.032992 (rank : 47) | θ value | 1.38821 (rank : 16) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 576 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P11679, Q3KQK5, Q3TGI1, Q3TJE1, Q3TKY7, Q61463, Q61518, Q61519 | Gene names | Krt8, Krt2-8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8) (Cytokeratin endo A). | |||||
|
K2C8_HUMAN
|
||||||
| NC score | 0.032979 (rank : 48) | θ value | 2.36792 (rank : 26) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 593 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P05787, Q14099, Q14716, Q14717, Q53GJ0, Q6DHW5, Q6GMY0, Q96J60 | Gene names | KRT8, CYK8 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cytoskeletal 8 (Cytokeratin-8) (CK-8) (Keratin-8) (K8). | |||||
|
MYO5A_MOUSE
|
||||||
| NC score | 0.032032 (rank : 49) | θ value | 5.27518 (rank : 42) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
LAMA4_HUMAN
|
||||||
| NC score | 0.031955 (rank : 50) | θ value | 5.27518 (rank : 41) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 708 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q16363, Q14735, Q15335, Q4LE44, Q5SZG8, Q9UE18, Q9UJN9 | Gene names | LAMA4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
SLK_HUMAN
|
||||||
| NC score | 0.031447 (rank : 51) | θ value | 0.163984 (rank : 7) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1775 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H2G2, O00211, Q6P1Z4, Q86WU7, Q86WW1, Q92603, Q9NQL0, Q9NQL1 | Gene names | SLK, KIAA0204, STK2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (hSLK) (Serine/threonine-protein kinase 2) (CTCL tumor antigen se20-9). | |||||
|
ROCK1_MOUSE
|
||||||
| NC score | 0.030628 (rank : 52) | θ value | 0.813845 (rank : 14) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 2126 | Shared Neighborhood Hits | 40 | |
| SwissProt Accessions | P70335, Q8C3G4, Q8C7H0 | Gene names | Rock1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK). | |||||
|
SLK_MOUSE
|
||||||
| NC score | 0.030225 (rank : 53) | θ value | 0.279714 (rank : 8) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1798 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O54988, Q80U65, Q8CAU2, Q8CDW2, Q9WU41 | Gene names | Slk, Kiaa0204, Stk2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STE20-like serine/threonine-protein kinase (EC 2.7.11.1) (STE20-like kinase) (STE20-related serine/threonine-protein kinase) (STE20-related kinase) (mSLK) (Serine/threonine-protein kinase 2) (STE20-related kinase SMAK) (Etk4). | |||||
|
SAPS1_HUMAN
|
||||||
| NC score | 0.030127 (rank : 54) | θ value | 0.47712 (rank : 12) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UPN7, Q504V2, Q6NVJ6, Q9BU97 | Gene names | SAPS1, KIAA1115 | |||
|
Domain Architecture |

|
|||||
| Description | SAPS domain family member 1. | |||||
|
CTTB2_HUMAN
|
||||||
| NC score | 0.029834 (rank : 55) | θ value | 5.27518 (rank : 39) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 1061 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8WZ74, O43389, Q7LG11, Q9C0A5 | Gene names | CTTNBP2, C7orf8, CORTBP2, KIAA1758 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cortactin-binding protein 2 (CortBP2). | |||||
|
TRIM2_MOUSE
|
||||||
| NC score | 0.012340 (rank : 56) | θ value | 8.99809 (rank : 48) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9ESN6, Q3UHP5, Q8C981 | Gene names | Trim2, Narf | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 2 (Neural activity-related RING finger protein). | |||||
|
TRIM2_HUMAN
|
||||||
| NC score | 0.012301 (rank : 57) | θ value | 8.99809 (rank : 47) | |||
| Query Neighborhood Hits | 48 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9C040, O60272, Q9BSI9, Q9UFZ1 | Gene names | TRIM2, KIAA0517, RNF86 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 2 (RING finger protein 86). | |||||