Please be patient as the page loads
|
UAP1_HUMAN
|
||||||
| SwissProt Accessions | Q16222, Q5VTA9, Q5VTB0, Q5VTB1, Q96GM2 | Gene names | UAP1, SPAG2 | |||
|
Domain Architecture |
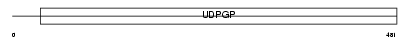
|
|||||
| Description | UDP-N-acetylhexosamine pyrophosphorylase (Antigen X) (AGX) (Sperm- associated antigen 2) [Includes: UDP-N-acetylgalactosamine pyrophosphorylase (EC 2.7.7.-) (AGX-1); UDP-N-acetylglucosamine pyrophosphorylase (EC 2.7.7.23) (AGX-2)]. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
UAP1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16222, Q5VTA9, Q5VTB0, Q5VTB1, Q96GM2 | Gene names | UAP1, SPAG2 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylhexosamine pyrophosphorylase (Antigen X) (AGX) (Sperm- associated antigen 2) [Includes: UDP-N-acetylgalactosamine pyrophosphorylase (EC 2.7.7.-) (AGX-1); UDP-N-acetylglucosamine pyrophosphorylase (EC 2.7.7.23) (AGX-2)]. | |||||
|
UAP1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.998916 (rank : 2) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91YN5, Q8BG76, Q8BXD6, Q8VD59 | Gene names | Uap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-N-acetylhexosamine pyrophosphorylase [Includes: UDP-N- acetylgalactosamine pyrophosphorylase (EC 2.7.7.-); UDP-N- acetylglucosamine pyrophosphorylase (EC 2.7.7.23)]. | |||||
|
UGPA2_MOUSE
|
||||||
| θ value | 1.38499e-08 (rank : 3) | NC score | 0.397393 (rank : 3) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZJ5, Q8R3D2 | Gene names | Ugp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UTP--glucose-1-phosphate uridylyltransferase 2 (EC 2.7.7.9) (UDP- glucose pyrophosphorylase 2) (UDPGP 2) (UGPase 2). | |||||
|
UGPA1_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 4) | NC score | 0.390457 (rank : 4) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07131 | Gene names | UGP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UTP--glucose-1-phosphate uridylyltransferase 1 (EC 2.7.7.9) (UDP- glucose pyrophosphorylase 1) (UDPGP 1) (UGPase 1). | |||||
|
UGPA2_HUMAN
|
||||||
| θ value | 5.26297e-08 (rank : 5) | NC score | 0.387057 (rank : 5) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16851, Q86Y81, Q9BU15 | Gene names | UGP2 | |||
|
Domain Architecture |
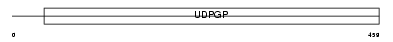
|
|||||
| Description | UTP--glucose-1-phosphate uridylyltransferase 2 (EC 2.7.7.9) (UDP- glucose pyrophosphorylase 2) (UDPGP 2) (UGPase 2). | |||||
|
TSCC1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 6) | NC score | 0.045904 (rank : 6) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K0G5 | Gene names | Tssc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TSSC1. | |||||
|
ADA20_HUMAN
|
||||||
| θ value | 4.03905 (rank : 7) | NC score | 0.013266 (rank : 7) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43506, Q9UKJ9 | Gene names | ADAM20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 20). | |||||
|
UAP1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16222, Q5VTA9, Q5VTB0, Q5VTB1, Q96GM2 | Gene names | UAP1, SPAG2 | |||
|
Domain Architecture |

|
|||||
| Description | UDP-N-acetylhexosamine pyrophosphorylase (Antigen X) (AGX) (Sperm- associated antigen 2) [Includes: UDP-N-acetylgalactosamine pyrophosphorylase (EC 2.7.7.-) (AGX-1); UDP-N-acetylglucosamine pyrophosphorylase (EC 2.7.7.23) (AGX-2)]. | |||||
|
UAP1_MOUSE
|
||||||
| NC score | 0.998916 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q91YN5, Q8BG76, Q8BXD6, Q8VD59 | Gene names | Uap1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UDP-N-acetylhexosamine pyrophosphorylase [Includes: UDP-N- acetylgalactosamine pyrophosphorylase (EC 2.7.7.-); UDP-N- acetylglucosamine pyrophosphorylase (EC 2.7.7.23)]. | |||||
|
UGPA2_MOUSE
|
||||||
| NC score | 0.397393 (rank : 3) | θ value | 1.38499e-08 (rank : 3) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZJ5, Q8R3D2 | Gene names | Ugp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UTP--glucose-1-phosphate uridylyltransferase 2 (EC 2.7.7.9) (UDP- glucose pyrophosphorylase 2) (UDPGP 2) (UGPase 2). | |||||
|
UGPA1_HUMAN
|
||||||
| NC score | 0.390457 (rank : 4) | θ value | 4.0297e-08 (rank : 4) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q07131 | Gene names | UGP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UTP--glucose-1-phosphate uridylyltransferase 1 (EC 2.7.7.9) (UDP- glucose pyrophosphorylase 1) (UDPGP 1) (UGPase 1). | |||||
|
UGPA2_HUMAN
|
||||||
| NC score | 0.387057 (rank : 5) | θ value | 5.26297e-08 (rank : 5) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q16851, Q86Y81, Q9BU15 | Gene names | UGP2 | |||
|
Domain Architecture |

|
|||||
| Description | UTP--glucose-1-phosphate uridylyltransferase 2 (EC 2.7.7.9) (UDP- glucose pyrophosphorylase 2) (UDPGP 2) (UGPase 2). | |||||
|
TSCC1_MOUSE
|
||||||
| NC score | 0.045904 (rank : 6) | θ value | 1.38821 (rank : 6) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K0G5 | Gene names | Tssc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein TSSC1. | |||||
|
ADA20_HUMAN
|
||||||
| NC score | 0.013266 (rank : 7) | θ value | 4.03905 (rank : 7) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 143 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43506, Q9UKJ9 | Gene names | ADAM20 | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 20 precursor (EC 3.4.24.-) (A disintegrin and metalloproteinase domain 20). | |||||