Please be patient as the page loads
|
TNMD_MOUSE
|
||||||
| SwissProt Accessions | Q9EP64, Q8CET4 | Gene names | Tnmd, Chm1l | |||
|
Domain Architecture |
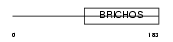
|
|||||
| Description | Tenomodulin (TeM) (mTeM) (Chondromodulin-I-like protein) (ChM1L) (mChM1L) (Myodulin) (Tendin). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
TNMD_MOUSE
|
||||||
| θ value | 9.82853e-156 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EP64, Q8CET4 | Gene names | Tnmd, Chm1l | |||
|
Domain Architecture |

|
|||||
| Description | Tenomodulin (TeM) (mTeM) (Chondromodulin-I-like protein) (ChM1L) (mChM1L) (Myodulin) (Tendin). | |||||
|
TNMD_HUMAN
|
||||||
| θ value | 2.12077e-150 (rank : 2) | NC score | 0.999084 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H2S6, Q9HBX0, Q9UJG0 | Gene names | TNMD, CHM1L | |||
|
Domain Architecture |
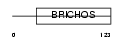
|
|||||
| Description | Tenomodulin (TeM) (hTeM) (Chondromodulin-I-like protein) (ChM1L) (hChM1L) (Myodulin) (Tendin). | |||||
|
LECT1_HUMAN
|
||||||
| θ value | 1.46268e-50 (rank : 3) | NC score | 0.897177 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75829, Q9UM18 | Gene names | LECT1, CHMI | |||
|
Domain Architecture |
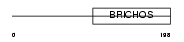
|
|||||
| Description | Chondromodulin-1 precursor (Chondromodulin-I) (ChM-I) (Leukocyte cell- derived chemotaxin 1) [Contains: Chondrosurfactant protein (CH-SP)]. | |||||
|
LECT1_MOUSE
|
||||||
| θ value | 1.91031e-50 (rank : 4) | NC score | 0.899413 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1F6, Q9CXU5 | Gene names | Lect1, Chmi | |||
|
Domain Architecture |
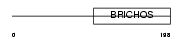
|
|||||
| Description | Chondromodulin-1 precursor (Chondromodulin-I) (ChM-I) (Leukocyte cell- derived chemotaxin 1) [Contains: Chondrosurfactant protein (CH-SP)]. | |||||
|
BLOT_HUMAN
|
||||||
| θ value | 0.0148317 (rank : 5) | NC score | 0.208164 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86XP6, Q6UWS6 | Gene names | BLOT, GDDR, TFIZ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Blottin precursor (Trefoil factor interactions(z) 1) (Down-regulated in gastric cancer). | |||||
|
AMYC_HUMAN
|
||||||
| θ value | 1.81305 (rank : 6) | NC score | 0.052686 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19961, Q9UBH3 | Gene names | AMY2B | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-amylase 2B precursor (EC 3.2.1.1) (1,4-alpha-D-glucan glucanohydrolase) (Alpha-amylase carcinoid). | |||||
|
AMYP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 7) | NC score | 0.052660 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P04746, Q9UBH3 | Gene names | AMY2A | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic alpha-amylase precursor (EC 3.2.1.1) (PA) (1,4-alpha-D- glucan glucanohydrolase). | |||||
|
AMYS_HUMAN
|
||||||
| θ value | 1.81305 (rank : 8) | NC score | 0.052711 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P04745, Q13763 | Gene names | AMY1A, AMY1 | |||
|
Domain Architecture |

|
|||||
| Description | Salivary alpha-amylase precursor (EC 3.2.1.1) (1,4-alpha-D-glucan glucanohydrolase). | |||||
|
BLOT_MOUSE
|
||||||
| θ value | 1.81305 (rank : 9) | NC score | 0.148702 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CQS6 | Gene names | Blot | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Blottin precursor. | |||||
|
MPP5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.026119 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLB2 | Gene names | Mpp5, Pals1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5 (Protein associated with Lin-7 1). | |||||
|
NTNG1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 11) | NC score | 0.026020 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2I2, Q5VU86, Q5VU87, Q5VU89, Q5VU90, Q5VU91, Q7Z2Y3, Q8N633 | Gene names | NTNG1, KIAA0976, LMNT1 | |||
|
Domain Architecture |
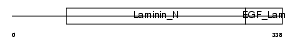
|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
NTNG1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 12) | NC score | 0.025989 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R4G0, Q68FE5, Q69ZU3, Q8R4F3, Q8R4F4, Q8R4F5, Q8R4F6, Q8R4F7, Q8R4F8, Q8R4F9, Q9ESR3, Q9ESR4, Q9ESR5, Q9ESR6, Q9ESR7, Q9ESR8 | Gene names | Ntng1, Kiaa0976, Lmnt1 | |||
|
Domain Architecture |
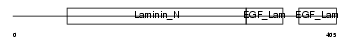
|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
DPOLB_MOUSE
|
||||||
| θ value | 5.27518 (rank : 13) | NC score | 0.024867 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K409, Q3UAB6, Q922Z7 | Gene names | Polb | |||
|
Domain Architecture |
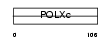
|
|||||
| Description | DNA polymerase beta (EC 2.7.7.7) (EC 4.2.99.-). | |||||
|
MPP5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 14) | NC score | 0.022650 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N3R9, Q7Z631, Q86T98, Q8N7I5, Q9H9Q0 | Gene names | MPP5 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5. | |||||
|
RFWD2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 15) | NC score | 0.018467 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NHY2, Q6H103, Q9H6L7 | Gene names | RFWD2, COP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (hCOP1). | |||||
|
CU005_HUMAN
|
||||||
| θ value | 6.88961 (rank : 16) | NC score | 0.021477 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3R5, Q9UEZ3 | Gene names | C21orf5, KIAA0933 | |||
|
Domain Architecture |
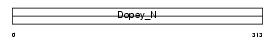
|
|||||
| Description | Protein C21orf5. | |||||
|
NTNG2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 17) | NC score | 0.020006 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96CW9, Q6UXY0, Q96JH0 | Gene names | NTNG2, KIAA1857, LMNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G2 precursor (Laminet-2). | |||||
|
RFWD2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 18) | NC score | 0.015701 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R1A8 | Gene names | Rfwd2, Cop1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (mCOP1). | |||||
|
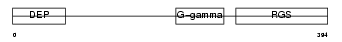
RGS7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 19) | NC score | 0.006396 (rank : 22) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49802, Q8TD66, Q8TD67, Q9UNU7, Q9Y6B9 | Gene names | RGS7 | |||
|
Domain Architecture |
|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
TFE2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 20) | NC score | 0.013395 (rank : 21) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
PSPC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 21) | NC score | 0.060950 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11686, P11687, Q12793, Q7Z5D0 | Gene names | SFTPC, SFTP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein C precursor (SP-C) (SP5) (Pulmonary surfactant-associated proteolipid SPL(Val)). | |||||
|
PSPC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 22) | NC score | 0.104146 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21841 | Gene names | Sftpc, Sftp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein C precursor (SP-C) (SP5) (Pulmonary surfactant-associated proteolipid SPL(Val)). | |||||
|
TNMD_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 9.82853e-156 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9EP64, Q8CET4 | Gene names | Tnmd, Chm1l | |||
|
Domain Architecture |

|
|||||
| Description | Tenomodulin (TeM) (mTeM) (Chondromodulin-I-like protein) (ChM1L) (mChM1L) (Myodulin) (Tendin). | |||||
|
TNMD_HUMAN
|
||||||
| NC score | 0.999084 (rank : 2) | θ value | 2.12077e-150 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9H2S6, Q9HBX0, Q9UJG0 | Gene names | TNMD, CHM1L | |||
|
Domain Architecture |

|
|||||
| Description | Tenomodulin (TeM) (hTeM) (Chondromodulin-I-like protein) (ChM1L) (hChM1L) (Myodulin) (Tendin). | |||||
|
LECT1_MOUSE
|
||||||
| NC score | 0.899413 (rank : 3) | θ value | 1.91031e-50 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9Z1F6, Q9CXU5 | Gene names | Lect1, Chmi | |||
|
Domain Architecture |

|
|||||
| Description | Chondromodulin-1 precursor (Chondromodulin-I) (ChM-I) (Leukocyte cell- derived chemotaxin 1) [Contains: Chondrosurfactant protein (CH-SP)]. | |||||
|
LECT1_HUMAN
|
||||||
| NC score | 0.897177 (rank : 4) | θ value | 1.46268e-50 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75829, Q9UM18 | Gene names | LECT1, CHMI | |||
|
Domain Architecture |

|
|||||
| Description | Chondromodulin-1 precursor (Chondromodulin-I) (ChM-I) (Leukocyte cell- derived chemotaxin 1) [Contains: Chondrosurfactant protein (CH-SP)]. | |||||
|
BLOT_HUMAN
|
||||||
| NC score | 0.208164 (rank : 5) | θ value | 0.0148317 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q86XP6, Q6UWS6 | Gene names | BLOT, GDDR, TFIZ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Blottin precursor (Trefoil factor interactions(z) 1) (Down-regulated in gastric cancer). | |||||
|
BLOT_MOUSE
|
||||||
| NC score | 0.148702 (rank : 6) | θ value | 1.81305 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9CQS6 | Gene names | Blot | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Blottin precursor. | |||||
|
PSPC_MOUSE
|
||||||
| NC score | 0.104146 (rank : 7) | θ value | θ > 10 (rank : 22) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21841 | Gene names | Sftpc, Sftp2 | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein C precursor (SP-C) (SP5) (Pulmonary surfactant-associated proteolipid SPL(Val)). | |||||
|
PSPC_HUMAN
|
||||||
| NC score | 0.060950 (rank : 8) | θ value | θ > 10 (rank : 21) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11686, P11687, Q12793, Q7Z5D0 | Gene names | SFTPC, SFTP2 | |||
|
Domain Architecture |

|
|||||
| Description | Pulmonary surfactant-associated protein C precursor (SP-C) (SP5) (Pulmonary surfactant-associated proteolipid SPL(Val)). | |||||
|
AMYS_HUMAN
|
||||||
| NC score | 0.052711 (rank : 9) | θ value | 1.81305 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P04745, Q13763 | Gene names | AMY1A, AMY1 | |||
|
Domain Architecture |

|
|||||
| Description | Salivary alpha-amylase precursor (EC 3.2.1.1) (1,4-alpha-D-glucan glucanohydrolase). | |||||
|
AMYC_HUMAN
|
||||||
| NC score | 0.052686 (rank : 10) | θ value | 1.81305 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19961, Q9UBH3 | Gene names | AMY2B | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-amylase 2B precursor (EC 3.2.1.1) (1,4-alpha-D-glucan glucanohydrolase) (Alpha-amylase carcinoid). | |||||
|
AMYP_HUMAN
|
||||||
| NC score | 0.052660 (rank : 11) | θ value | 1.81305 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P04746, Q9UBH3 | Gene names | AMY2A | |||
|
Domain Architecture |

|
|||||
| Description | Pancreatic alpha-amylase precursor (EC 3.2.1.1) (PA) (1,4-alpha-D- glucan glucanohydrolase). | |||||
|
MPP5_MOUSE
|
||||||
| NC score | 0.026119 (rank : 12) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JLB2 | Gene names | Mpp5, Pals1 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5 (Protein associated with Lin-7 1). | |||||
|
NTNG1_HUMAN
|
||||||
| NC score | 0.026020 (rank : 13) | θ value | 2.36792 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2I2, Q5VU86, Q5VU87, Q5VU89, Q5VU90, Q5VU91, Q7Z2Y3, Q8N633 | Gene names | NTNG1, KIAA0976, LMNT1 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
NTNG1_MOUSE
|
||||||
| NC score | 0.025989 (rank : 14) | θ value | 2.36792 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 246 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8R4G0, Q68FE5, Q69ZU3, Q8R4F3, Q8R4F4, Q8R4F5, Q8R4F6, Q8R4F7, Q8R4F8, Q8R4F9, Q9ESR3, Q9ESR4, Q9ESR5, Q9ESR6, Q9ESR7, Q9ESR8 | Gene names | Ntng1, Kiaa0976, Lmnt1 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G1 precursor (Laminet-1). | |||||
|
DPOLB_MOUSE
|
||||||
| NC score | 0.024867 (rank : 15) | θ value | 5.27518 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8K409, Q3UAB6, Q922Z7 | Gene names | Polb | |||
|
Domain Architecture |

|
|||||
| Description | DNA polymerase beta (EC 2.7.7.7) (EC 4.2.99.-). | |||||
|
MPP5_HUMAN
|
||||||
| NC score | 0.022650 (rank : 16) | θ value | 5.27518 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N3R9, Q7Z631, Q86T98, Q8N7I5, Q9H9Q0 | Gene names | MPP5 | |||
|
Domain Architecture |

|
|||||
| Description | MAGUK p55 subfamily member 5. | |||||
|
CU005_HUMAN
|
||||||
| NC score | 0.021477 (rank : 17) | θ value | 6.88961 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 42 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9Y3R5, Q9UEZ3 | Gene names | C21orf5, KIAA0933 | |||
|
Domain Architecture |
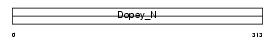
|
|||||
| Description | Protein C21orf5. | |||||
|
NTNG2_HUMAN
|
||||||
| NC score | 0.020006 (rank : 18) | θ value | 6.88961 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96CW9, Q6UXY0, Q96JH0 | Gene names | NTNG2, KIAA1857, LMNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Netrin G2 precursor (Laminet-2). | |||||
|
RFWD2_HUMAN
|
||||||
| NC score | 0.018467 (rank : 19) | θ value | 5.27518 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 518 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NHY2, Q6H103, Q9H6L7 | Gene names | RFWD2, COP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (hCOP1). | |||||
|
RFWD2_MOUSE
|
||||||
| NC score | 0.015701 (rank : 20) | θ value | 8.99809 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 535 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R1A8 | Gene names | Rfwd2, Cop1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger and WD repeat domain protein 2 (EC 6.3.2.-) (Ubiquitin- protein ligase COP1) (Constitutive photomorphogenesis protein 1 homolog) (mCOP1). | |||||
|
TFE2_MOUSE
|
||||||
| NC score | 0.013395 (rank : 21) | θ value | 8.99809 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 118 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P15806, Q3U153, Q8CAH9, Q8VCY4, Q922S2, Q99MB8, Q9CYJ4 | Gene names | Tcf3, Alf2, Me2, Tcfe2a | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription factor E2-alpha (Immunoglobulin enhancer-binding factor E12/E47) (Transcription factor 3) (TCF-3) (Transcription factor A1). | |||||
|
RGS7_HUMAN
|
||||||
| NC score | 0.006396 (rank : 22) | θ value | 8.99809 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49802, Q8TD66, Q8TD67, Q9UNU7, Q9Y6B9 | Gene names | RGS7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||