Please be patient as the page loads
|
THTM_HUMAN
|
||||||
| SwissProt Accessions | P25325, O75750 | Gene names | MPST, TST2 | |||
|
Domain Architecture |
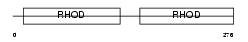
|
|||||
| Description | 3-mercaptopyruvate sulfurtransferase (EC 2.8.1.2) (MST). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
THTM_HUMAN
|
||||||
| θ value | 5.9214e-177 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P25325, O75750 | Gene names | MPST, TST2 | |||
|
Domain Architecture |

|
|||||
| Description | 3-mercaptopyruvate sulfurtransferase (EC 2.8.1.2) (MST). | |||||
|
THTM_MOUSE
|
||||||
| θ value | 1.46868e-151 (rank : 2) | NC score | 0.998062 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99J99 | Gene names | Mpst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-mercaptopyruvate sulfurtransferase (EC 2.8.1.2) (MST). | |||||
|
THTR_MOUSE
|
||||||
| θ value | 2.07809e-105 (rank : 3) | NC score | 0.985778 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52196 | Gene names | Tst | |||
|
Domain Architecture |
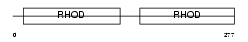
|
|||||
| Description | Thiosulfate sulfurtransferase (EC 2.8.1.1) (Rhodanese). | |||||
|
THTR_HUMAN
|
||||||
| θ value | 1.75921e-104 (rank : 4) | NC score | 0.985586 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16762 | Gene names | TST | |||
|
Domain Architecture |
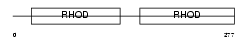
|
|||||
| Description | Thiosulfate sulfurtransferase (EC 2.8.1.1) (Rhodanese). | |||||
|
KAT_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 5) | NC score | 0.175764 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFU3, Q5SY48, Q5SY49, Q5SY50, Q5SY51, Q8NFU2, Q9BV22 | Gene names | KAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative thiosulfate sulfurtransferase KAT (EC 2.8.1.1). | |||||
|
ZN167_HUMAN
|
||||||
| θ value | 1.06291 (rank : 6) | NC score | 0.001488 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P0L1, Q6WL09, Q96FQ2 | Gene names | ZNF167, ZNF448, ZNF64 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 167 (Zinc finger protein 64) (Zinc finger protein 448) (ZFP). | |||||
|
FINC_MOUSE
|
||||||
| θ value | 1.81305 (rank : 7) | NC score | 0.017831 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
FINC_HUMAN
|
||||||
| θ value | 3.0926 (rank : 8) | NC score | 0.016814 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
SDHL_HUMAN
|
||||||
| θ value | 4.03905 (rank : 9) | NC score | 0.035614 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20132 | Gene names | SDS, SDH | |||
|
Domain Architecture |
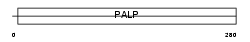
|
|||||
| Description | L-serine dehydratase (EC 4.3.1.17) (L-serine deaminase). | |||||
|
PPM1F_HUMAN
|
||||||
| θ value | 5.27518 (rank : 10) | NC score | 0.026726 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49593, Q96PM2 | Gene names | PPM1F, KIAA0015, POPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Partner of PIX 2) (hFEM-2) (Protein phosphatase 1F). | |||||
|
RAB5B_HUMAN
|
||||||
| θ value | 6.88961 (rank : 11) | NC score | 0.003898 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61020, P35239, P35277, Q6PIK9, Q86TH0, Q8IXL2 | Gene names | RAB5B | |||
|
Domain Architecture |
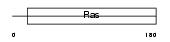
|
|||||
| Description | Ras-related protein Rab-5B. | |||||
|
RAB5B_MOUSE
|
||||||
| θ value | 6.88961 (rank : 12) | NC score | 0.003898 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61021, P35239, P35277 | Gene names | Rab5b | |||
|
Domain Architecture |
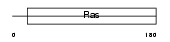
|
|||||
| Description | Ras-related protein Rab-5B. | |||||
|
THTM_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 5.9214e-177 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P25325, O75750 | Gene names | MPST, TST2 | |||
|
Domain Architecture |

|
|||||
| Description | 3-mercaptopyruvate sulfurtransferase (EC 2.8.1.2) (MST). | |||||
|
THTM_MOUSE
|
||||||
| NC score | 0.998062 (rank : 2) | θ value | 1.46868e-151 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q99J99 | Gene names | Mpst | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 3-mercaptopyruvate sulfurtransferase (EC 2.8.1.2) (MST). | |||||
|
THTR_MOUSE
|
||||||
| NC score | 0.985778 (rank : 3) | θ value | 2.07809e-105 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P52196 | Gene names | Tst | |||
|
Domain Architecture |

|
|||||
| Description | Thiosulfate sulfurtransferase (EC 2.8.1.1) (Rhodanese). | |||||
|
THTR_HUMAN
|
||||||
| NC score | 0.985586 (rank : 4) | θ value | 1.75921e-104 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16762 | Gene names | TST | |||
|
Domain Architecture |

|
|||||
| Description | Thiosulfate sulfurtransferase (EC 2.8.1.1) (Rhodanese). | |||||
|
KAT_HUMAN
|
||||||
| NC score | 0.175764 (rank : 5) | θ value | 0.0961366 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8NFU3, Q5SY48, Q5SY49, Q5SY50, Q5SY51, Q8NFU2, Q9BV22 | Gene names | KAT | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Putative thiosulfate sulfurtransferase KAT (EC 2.8.1.1). | |||||
|
SDHL_HUMAN
|
||||||
| NC score | 0.035614 (rank : 6) | θ value | 4.03905 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P20132 | Gene names | SDS, SDH | |||
|
Domain Architecture |

|
|||||
| Description | L-serine dehydratase (EC 4.3.1.17) (L-serine deaminase). | |||||
|
PPM1F_HUMAN
|
||||||
| NC score | 0.026726 (rank : 7) | θ value | 5.27518 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P49593, Q96PM2 | Gene names | PPM1F, KIAA0015, POPX2 | |||
|
Domain Architecture |

|
|||||
| Description | Ca(2+)/calmodulin-dependent protein kinase phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase) (CaMKPase) (Partner of PIX 2) (hFEM-2) (Protein phosphatase 1F). | |||||
|
FINC_MOUSE
|
||||||
| NC score | 0.017831 (rank : 8) | θ value | 1.81305 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P11276, Q61567, Q61568, Q61569, Q64233, Q80UI4 | Gene names | Fn1 | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN). | |||||
|
FINC_HUMAN
|
||||||
| NC score | 0.016814 (rank : 9) | θ value | 3.0926 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P02751, O95609, O95610, Q14312, Q14325, Q14326, Q6LDP6, Q86T27, Q8IVI8, Q96KP7, Q96KP8, Q96KP9, Q9H1B8, Q9HAP3, Q9UMK2 | Gene names | FN1, FN | |||
|
Domain Architecture |

|
|||||
| Description | Fibronectin precursor (FN) (Cold-insoluble globulin) (CIG). | |||||
|
RAB5B_HUMAN
|
||||||
| NC score | 0.003898 (rank : 10) | θ value | 6.88961 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61020, P35239, P35277, Q6PIK9, Q86TH0, Q8IXL2 | Gene names | RAB5B | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-5B. | |||||
|
RAB5B_MOUSE
|
||||||
| NC score | 0.003898 (rank : 11) | θ value | 6.88961 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61021, P35239, P35277 | Gene names | Rab5b | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-5B. | |||||
|
ZN167_HUMAN
|
||||||
| NC score | 0.001488 (rank : 12) | θ value | 1.06291 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9P0L1, Q6WL09, Q96FQ2 | Gene names | ZNF167, ZNF448, ZNF64 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 167 (Zinc finger protein 64) (Zinc finger protein 448) (ZFP). | |||||