Please be patient as the page loads
|
SMRD1_HUMAN
|
||||||
| SwissProt Accessions | Q96GM5, Q92924, Q9Y635 | Gene names | SMARCD1, BAF60A | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 (SWI/SNF complex 60 kDa subunit) (60 kDa BRG-1/Brm-associated factor subunit A) (BRG1-associated factor 60A). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SMRD1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q96GM5, Q92924, Q9Y635 | Gene names | SMARCD1, BAF60A | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 (SWI/SNF complex 60 kDa subunit) (60 kDa BRG-1/Brm-associated factor subunit A) (BRG1-associated factor 60A). | |||||
|
SMRD1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999927 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q61466, P70384, Q8R0I7 | Gene names | Smarcd1, Baf60a, D15Kz1 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 (SWI/SNF complex 60 kDa subunit) (60 kDa BRG-1/Brm-associated factor subunit A) (BRG1-associated factor 60A) (D15KZ1 protein). | |||||
|
SMRD3_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.982515 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6STE5, Q75MJ2, Q75MR8, Q92926, Q9BUH1 | Gene names | SMARCD3, BAF60C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 3 (60 kDa BRG-1/Brm-associated factor subunit C) (BRG1-associated factor 60C). | |||||
|
SMRD3_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.982824 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P9Z1, Q921D3, Q9CX89, Q9DB99 | Gene names | Smarcd3, Baf60c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 3 (60 kDa BRG-1/Brm-associated factor subunit C) (BRG1-associated factor 60C) (mBAF60c). | |||||
|
SMRD2_MOUSE
|
||||||
| θ value | 5.93515e-169 (rank : 5) | NC score | 0.983476 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99JR8 | Gene names | Smarcd2, Baf60b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 2 (60 kDa BRG-1/Brm-associated factor subunit B) (BRG1-associated factor 60B). | |||||
|
SMRD2_HUMAN
|
||||||
| θ value | 1.32529e-160 (rank : 6) | NC score | 0.982013 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92925, Q7L2I6, Q9UHZ1 | Gene names | SMARCD2, BAF60B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 2 (60 kDa BRG-1/Brm-associated factor subunit B) (BRG1-associated factor 60B). | |||||
|
SMCA4_HUMAN
|
||||||
| θ value | 0.00102713 (rank : 7) | NC score | 0.049255 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
SMCA2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 8) | NC score | 0.043037 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
SFPQ_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 9) | NC score | 0.044785 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
TFG_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 10) | NC score | 0.095196 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92734, Q15656 | Gene names | TFG | |||
|
Domain Architecture |

|
|||||
| Description | Protein TFG (TRK-fused gene protein). | |||||
|
FMN2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.042762 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
ZN207_HUMAN
|
||||||
| θ value | 0.21417 (rank : 12) | NC score | 0.066885 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
FOG1_HUMAN
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.011431 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
SF3B4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 14) | NC score | 0.031445 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
ARI1B_HUMAN
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.040317 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
RBM12_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.049373 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
RBM12_MOUSE
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.048019 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
SFPQ_HUMAN
|
||||||
| θ value | 0.47712 (rank : 18) | NC score | 0.036105 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
HCN2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.018070 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
RSMB_HUMAN
|
||||||
| θ value | 0.62314 (rank : 20) | NC score | 0.068663 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P14678, Q15490, Q9UIS5 | Gene names | SNRPB, COD, SNRPB1 | |||
|
Domain Architecture |
|
|||||
| Description | Small nuclear ribonucleoprotein-associated proteins B and B' (snRNP-B) (Sm protein B/B') (Sm-B/Sm-B') (SmB/SmB'). | |||||
|
RSMB_MOUSE
|
||||||
| θ value | 0.62314 (rank : 21) | NC score | 0.069066 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P27048, Q3UJT1 | Gene names | Snrpb | |||
|
Domain Architecture |
|
|||||
| Description | Small nuclear ribonucleoprotein-associated protein B (snRNP-B) (Sm protein B) (Sm-B) (SmB). | |||||
|
WBP11_HUMAN
|
||||||
| θ value | 0.62314 (rank : 22) | NC score | 0.050073 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | 0.62314 (rank : 23) | NC score | 0.050698 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
LEG3_MOUSE
|
||||||
| θ value | 0.813845 (rank : 24) | NC score | 0.040288 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16110 | Gene names | Lgals3 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-34 galactoside-binding lectin). | |||||
|
PRPC_HUMAN
|
||||||
| θ value | 0.813845 (rank : 25) | NC score | 0.061914 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
ADIP_MOUSE
|
||||||
| θ value | 1.06291 (rank : 26) | NC score | 0.025436 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VC66, Q8BG59, Q8C7X0, Q8K2F7 | Gene names | Ssx2ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Afadin- and alpha-actinin-binding protein (ADIP) (Afadin DIL domain- interacting protein). | |||||
|
CO4A1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 27) | NC score | 0.021594 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
ELN_HUMAN
|
||||||
| θ value | 1.06291 (rank : 28) | NC score | 0.059641 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P15502, O15336, O15337, Q14233, Q14234, Q14235, Q14238, Q6P0L4, Q6ZWJ6, Q75MU5, Q7Z316, Q7Z3F5, Q9UMF5 | Gene names | ELN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
FMN2_MOUSE
|
||||||
| θ value | 1.06291 (rank : 29) | NC score | 0.037688 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
CCD93_HUMAN
|
||||||
| θ value | 1.38821 (rank : 30) | NC score | 0.018513 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q567U6, Q4LE78, Q53TJ2, Q8TBX5, Q9H6R5, Q9NV15 | Gene names | CCDC93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 93. | |||||
|
RU1C_HUMAN
|
||||||
| θ value | 1.38821 (rank : 31) | NC score | 0.065653 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P09234 | Gene names | SNRPC | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
RU1C_MOUSE
|
||||||
| θ value | 1.38821 (rank : 32) | NC score | 0.065154 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62241 | Gene names | Snrp1c, Snrpc | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
ANXA7_HUMAN
|
||||||
| θ value | 1.81305 (rank : 33) | NC score | 0.018229 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P20073 | Gene names | ANXA7, ANX7, SNX | |||
|
Domain Architecture |
|
|||||
| Description | Annexin A7 (Annexin VII) (Synexin). | |||||
|
LEG3_HUMAN
|
||||||
| θ value | 1.81305 (rank : 34) | NC score | 0.035365 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P17931, Q16005, Q96J47 | Gene names | LGALS3, MAC2 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-31) (Galactoside-binding protein) (GALBP). | |||||
|
SF3A2_MOUSE
|
||||||
| θ value | 1.81305 (rank : 35) | NC score | 0.034852 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
CO9A3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.018782 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
CS007_HUMAN
|
||||||
| θ value | 2.36792 (rank : 37) | NC score | 0.027350 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
SNRPA_MOUSE
|
||||||
| θ value | 2.36792 (rank : 38) | NC score | 0.028331 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62189, Q52LA2 | Gene names | Snrpa, Rnu1a-1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein A (U1 snRNP protein A) (U1A protein) (U1-A). | |||||
|
ANX11_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.016336 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
CN032_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.029998 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
FMNL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.025082 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.006812 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
NEUR4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.020657 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WWR8, Q96D64 | Gene names | NEU4 | |||
|
Domain Architecture |
|
|||||
| Description | Sialidase-4 (EC 3.2.1.18) (N-acetyl-alpha-neuraminidase 4). | |||||
|
PEF1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.019348 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BFY6, Q8VCT5, Q9CYW8, Q9D934 | Gene names | Pef1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
PRP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.021864 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
PYGO2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.034946 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
ATN1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.023431 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
BSN_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.025453 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
CO4A3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.017641 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
COIA1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.019429 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
DAAM1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 51) | NC score | 0.021202 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y4D1, Q86U34, Q8N1Z8, Q8TB39 | Gene names | DAAM1, KIAA0666 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
EMID2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 52) | NC score | 0.020965 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96A83 | Gene names | EMID2, COL26A1, EMU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXVI) chain precursor (EMI domain-containing protein 2) (Protein Emu2) (Emilin and multimerin domain-containing protein 2). | |||||
|
LTBP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 53) | NC score | 0.005292 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
MISS_MOUSE
|
||||||
| θ value | 4.03905 (rank : 54) | NC score | 0.049773 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
MTSS1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 55) | NC score | 0.035850 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
STRN3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.008647 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERG2 | Gene names | Strn3, Gs2na, Sg2na | |||
|
Domain Architecture |

|
|||||
| Description | Striatin-3 (Cell-cycle autoantigen SG2NA) (S/G2 antigen). | |||||
|
TMBI1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.022180 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q969X1, Q8N1R3, Q8TAM3, Q96K13 | Gene names | TMBIM1, RECS1 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane BAX inhibitor motif-containing protein 1 (RECS1 protein homolog). | |||||
|
CCNK_MOUSE
|
||||||
| θ value | 5.27518 (rank : 58) | NC score | 0.014728 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CS007_MOUSE
|
||||||
| θ value | 5.27518 (rank : 59) | NC score | 0.022597 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
BSN_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.026313 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
CO9A2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.014473 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07643 | Gene names | Col9a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
CPSF6_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.021822 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CPSF6_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.021200 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
FOSB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.013538 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.003683 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
LHX5_MOUSE
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.003716 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61375, P50459 | Gene names | Lhx5 | |||
|
Domain Architecture |
|
|||||
| Description | LIM/homeobox protein Lhx5. | |||||
|
RERE_MOUSE
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.014407 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
SF3A2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.025630 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SSBP3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.020629 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |
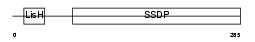
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.020629 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |
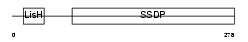
|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
CADH4_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.002852 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55283, Q2M208, Q5VZ44, Q9BZ05 | Gene names | CDH4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
CADH4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.002848 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P39038 | Gene names | Cdh4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
CN140_MOUSE
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.015743 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CCG1, Q8CCM8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf140 homolog. | |||||
|
COBA2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.012130 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P13942, Q07751, Q13271, Q13272, Q13273, Q99866, Q9UIP9 | Gene names | COL11A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
GCNL2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.012714 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
GSCR1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.013067 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
IRX3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.008972 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78415, Q7Z4A4, Q7Z4A5, Q8IVC6 | Gene names | IRX3, IRXB1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
IRX3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.009139 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P81067 | Gene names | Irx3, Irxb1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
KISS1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.023720 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15726, Q9HBP1 | Gene names | KISS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis-suppressor KiSS-1 precursor (Kisspeptin-1) [Contains: Metastin (Kisspeptin-54); Kisspeptin-14; Kisspeptin-13; Kisspeptin- 10]. | |||||
|
PRP1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.012614 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
RGS7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.008490 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49802, Q8TD66, Q8TD67, Q9UNU7, Q9Y6B9 | Gene names | RGS7 | |||
|
Domain Architecture |
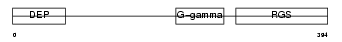
|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS7_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.008508 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54829 | Gene names | Rgs7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RSMN_HUMAN
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.038390 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P63162, P14648, P17135 | Gene names | SNRPN, HCERN3, SMN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
RSMN_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.038390 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P63163, P14648, P17135 | Gene names | Snrpn, Smn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
TMPS6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.001387 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IU80, Q5TI06, Q6UXD8, Q8IUE2, Q8IXV8 | Gene names | TMPRSS6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
SMRD1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q96GM5, Q92924, Q9Y635 | Gene names | SMARCD1, BAF60A | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 (SWI/SNF complex 60 kDa subunit) (60 kDa BRG-1/Brm-associated factor subunit A) (BRG1-associated factor 60A). | |||||
|
SMRD1_MOUSE
|
||||||
| NC score | 0.999927 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | Q61466, P70384, Q8R0I7 | Gene names | Smarcd1, Baf60a, D15Kz1 | |||
|
Domain Architecture |

|
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 (SWI/SNF complex 60 kDa subunit) (60 kDa BRG-1/Brm-associated factor subunit A) (BRG1-associated factor 60A) (D15KZ1 protein). | |||||
|
SMRD2_MOUSE
|
||||||
| NC score | 0.983476 (rank : 3) | θ value | 5.93515e-169 (rank : 5) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q99JR8 | Gene names | Smarcd2, Baf60b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 2 (60 kDa BRG-1/Brm-associated factor subunit B) (BRG1-associated factor 60B). | |||||
|
SMRD3_MOUSE
|
||||||
| NC score | 0.982824 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 31 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6P9Z1, Q921D3, Q9CX89, Q9DB99 | Gene names | Smarcd3, Baf60c | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 3 (60 kDa BRG-1/Brm-associated factor subunit C) (BRG1-associated factor 60C) (mBAF60c). | |||||
|
SMRD3_HUMAN
|
||||||
| NC score | 0.982515 (rank : 5) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 30 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q6STE5, Q75MJ2, Q75MR8, Q92926, Q9BUH1 | Gene names | SMARCD3, BAF60C | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 3 (60 kDa BRG-1/Brm-associated factor subunit C) (BRG1-associated factor 60C). | |||||
|
SMRD2_HUMAN
|
||||||
| NC score | 0.982013 (rank : 6) | θ value | 1.32529e-160 (rank : 6) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q92925, Q7L2I6, Q9UHZ1 | Gene names | SMARCD2, BAF60B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily D member 2 (60 kDa BRG-1/Brm-associated factor subunit B) (BRG1-associated factor 60B). | |||||
|
TFG_HUMAN
|
||||||
| NC score | 0.095196 (rank : 7) | θ value | 0.0563607 (rank : 10) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q92734, Q15656 | Gene names | TFG | |||
|
Domain Architecture |

|
|||||
| Description | Protein TFG (TRK-fused gene protein). | |||||
|
RSMB_MOUSE
|
||||||
| NC score | 0.069066 (rank : 8) | θ value | 0.62314 (rank : 21) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P27048, Q3UJT1 | Gene names | Snrpb | |||
|
Domain Architecture |

|
|||||
| Description | Small nuclear ribonucleoprotein-associated protein B (snRNP-B) (Sm protein B) (Sm-B) (SmB). | |||||
|
RSMB_HUMAN
|
||||||
| NC score | 0.068663 (rank : 9) | θ value | 0.62314 (rank : 20) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P14678, Q15490, Q9UIS5 | Gene names | SNRPB, COD, SNRPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Small nuclear ribonucleoprotein-associated proteins B and B' (snRNP-B) (Sm protein B/B') (Sm-B/Sm-B') (SmB/SmB'). | |||||
|
ZN207_HUMAN
|
||||||
| NC score | 0.066885 (rank : 10) | θ value | 0.21417 (rank : 12) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 326 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O43670, Q96HW5, Q9BUQ7 | Gene names | ZNF207 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 207. | |||||
|
RU1C_HUMAN
|
||||||
| NC score | 0.065653 (rank : 11) | θ value | 1.38821 (rank : 31) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P09234 | Gene names | SNRPC | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
RU1C_MOUSE
|
||||||
| NC score | 0.065154 (rank : 12) | θ value | 1.38821 (rank : 32) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q62241 | Gene names | Snrp1c, Snrpc | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) (U1C protein) (U1-C). | |||||
|
PRPC_HUMAN
|
||||||
| NC score | 0.061914 (rank : 13) | θ value | 0.813845 (rank : 25) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 165 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P02810, Q4VBP2, Q53XA2, Q6P2F6 | Gene names | PRH1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Salivary acidic proline-rich phosphoprotein 1/2 precursor (PRP-1/PRP- 2) (Parotid proline-rich protein 1/2) (Pr1/Pr2) (Protein C) (Parotid acidic protein) (Pa) (Parotid isoelectric focusing variant protein) (PIF-S) (Parotid double-band protein) (Db-s) [Contains: Salivary acidic proline-rich phosphoprotein 1/2; Salivary acidic proline-rich phosphoprotein 3/4 (PRP-3/PRP-4) (Protein A) (PIF-F) (Db-F); Peptide P-C]. | |||||
|
ELN_HUMAN
|
||||||
| NC score | 0.059641 (rank : 14) | θ value | 1.06291 (rank : 28) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 85 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P15502, O15336, O15337, Q14233, Q14234, Q14235, Q14238, Q6P0L4, Q6ZWJ6, Q75MU5, Q7Z316, Q7Z3F5, Q9UMF5 | Gene names | ELN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Elastin precursor (Tropoelastin). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.050698 (rank : 15) | θ value | 0.62314 (rank : 23) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.050073 (rank : 16) | θ value | 0.62314 (rank : 22) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
MISS_MOUSE
|
||||||
| NC score | 0.049773 (rank : 17) | θ value | 4.03905 (rank : 54) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 444 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9D7G9, Q7M749 | Gene names | Mapk1ip1, Miss | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | MAPK-interacting and spindle-stabilizing protein (Mitogen-activated protein kinase 1-interacting protein 1). | |||||
|
RBM12_HUMAN
|
||||||
| NC score | 0.049373 (rank : 18) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NTZ6, O94865, Q8N3B1, Q9H196 | Gene names | RBM12, KIAA0765 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
SMCA4_HUMAN
|
||||||
| NC score | 0.049255 (rank : 19) | θ value | 0.00102713 (rank : 7) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 597 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P51532, O95052, Q9HBD3 | Gene names | SMARCA4, BRG1, SNF2B, SNF2L4 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L4 (EC 3.6.1.-) (ATP- dependent helicase SMARCA4) (SNF2-beta) (BRG-1 protein) (Mitotic growth and transcription activator) (Brahma protein homolog 1) (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 4). | |||||
|
RBM12_MOUSE
|
||||||
| NC score | 0.048019 (rank : 20) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 313 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8R4X3, Q8K373, Q8R302, Q9CS80 | Gene names | Rbm12 | |||
|
Domain Architecture |

|
|||||
| Description | RNA-binding protein 12 (RNA-binding motif protein 12) (SH3/WW domain anchor protein in the nucleus) (SWAN). | |||||
|
SFPQ_MOUSE
|
||||||
| NC score | 0.044785 (rank : 21) | θ value | 0.0431538 (rank : 9) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 958 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q8VIJ6, Q9ERW2 | Gene names | Sfpq | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit). | |||||
|
SMCA2_HUMAN
|
||||||
| NC score | 0.043037 (rank : 22) | θ value | 0.0193708 (rank : 8) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P51531 | Gene names | SMARCA2, BRM, SNF2A, SNF2L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable global transcription activator SNF2L2 (EC 3.6.1.-) (ATP- dependent helicase SMARCA2) (SNF2-alpha) (SWI/SNF-related matrix- associated actin-dependent regulator of chromatin subfamily A member 2) (hBRM). | |||||
|
FMN2_HUMAN
|
||||||
| NC score | 0.042762 (rank : 23) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 484 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9NZ56, Q59GF6, Q5VU37, Q9NZ55 | Gene names | FMN2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Formin-2. | |||||
|
ARI1B_HUMAN
|
||||||
| NC score | 0.040317 (rank : 24) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 567 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8NFD5, Q5JRD1, Q5VYC4, Q8IZY8, Q8TEV0, Q8TF02, Q99491, Q9ULI5 | Gene names | ARID1B, DAN15, KIAA1235, OSA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AT-rich interactive domain-containing protein 1B (ARID domain- containing protein 1B) (Osa homolog 2) (hOsa2) (p250R) (BRG1-binding protein hELD/OSA1) (BRG1-associated factor 250b) (BAF250B). | |||||
|
LEG3_MOUSE
|
||||||
| NC score | 0.040288 (rank : 25) | θ value | 0.813845 (rank : 24) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16110 | Gene names | Lgals3 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-34 galactoside-binding lectin). | |||||
|
RSMN_HUMAN
|
||||||
| NC score | 0.038390 (rank : 26) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P63162, P14648, P17135 | Gene names | SNRPN, HCERN3, SMN | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
RSMN_MOUSE
|
||||||
| NC score | 0.038390 (rank : 27) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P63163, P14648, P17135 | Gene names | Snrpn, Smn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Small nuclear ribonucleoprotein-associated protein N (snRNP-N) (Sm protein N) (Sm-N) (SmN) (Sm-D) (Tissue-specific-splicing protein). | |||||
|
FMN2_MOUSE
|
||||||
| NC score | 0.037688 (rank : 28) | θ value | 1.06291 (rank : 29) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q9JL04 | Gene names | Fmn2 | |||
|
Domain Architecture |

|
|||||
| Description | Formin-2. | |||||
|
SFPQ_HUMAN
|
||||||
| NC score | 0.036105 (rank : 29) | θ value | 0.47712 (rank : 18) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 986 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P23246, P30808 | Gene names | SFPQ, PSF | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, proline- and glutamine-rich (Polypyrimidine tract- binding protein-associated-splicing factor) (PTB-associated-splicing factor) (PSF) (DNA-binding p52/p100 complex, 100 kDa subunit) (100 kDa DNA-pairing protein) (hPOMp100). | |||||
|
MTSS1_MOUSE
|
||||||
| NC score | 0.035850 (rank : 30) | θ value | 4.03905 (rank : 55) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 592 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q8R1S4, Q8BMM3, Q99LB3 | Gene names | Mtss1, Mim | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis suppressor protein 1 (Missing in metastasis protein). | |||||
|
LEG3_HUMAN
|
||||||
| NC score | 0.035365 (rank : 31) | θ value | 1.81305 (rank : 34) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P17931, Q16005, Q96J47 | Gene names | LGALS3, MAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-31) (Galactoside-binding protein) (GALBP). | |||||
|
PYGO2_HUMAN
|
||||||
| NC score | 0.034946 (rank : 32) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 189 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9BRQ0, Q8WYZ4, Q96CY2 | Gene names | PYGO2 | |||
|
Domain Architecture |

|
|||||
| Description | Pygopus homolog 2. | |||||
|
SF3A2_MOUSE
|
||||||
| NC score | 0.034852 (rank : 33) | θ value | 1.81305 (rank : 35) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 247 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q62203 | Gene names | Sf3a2, Sap62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
SF3B4_HUMAN
|
||||||
| NC score | 0.031445 (rank : 34) | θ value | 0.365318 (rank : 14) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q15427 | Gene names | SF3B4, SAP49 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3B subunit 4 (Spliceosome-associated protein 49) (SAP 49) (SF3b50) (Pre-mRNA-splicing factor SF3b 49 kDa subunit). | |||||
|
CN032_HUMAN
|
||||||
| NC score | 0.029998 (rank : 35) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 267 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NDC0, Q96BG5 | Gene names | C14orf32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf32. | |||||
|
SNRPA_MOUSE
|
||||||
| NC score | 0.028331 (rank : 36) | θ value | 2.36792 (rank : 38) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q62189, Q52LA2 | Gene names | Snrpa, Rnu1a-1 | |||
|
Domain Architecture |

|
|||||
| Description | U1 small nuclear ribonucleoprotein A (U1 snRNP protein A) (U1A protein) (U1-A). | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.027350 (rank : 37) | θ value | 2.36792 (rank : 37) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
BSN_MOUSE
|
||||||
| NC score | 0.026313 (rank : 38) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 2057 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O88737, Q6ZQB5 | Gene names | Bsn, Kiaa0434 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon. | |||||
|
SF3A2_HUMAN
|
||||||
| NC score | 0.025630 (rank : 39) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 263 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q15428, O75245 | Gene names | SF3A2, SAP62 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor 3A subunit 2 (Spliceosome-associated protein 62) (SAP 62) (SF3a66). | |||||
|
BSN_HUMAN
|
||||||
| NC score | 0.025453 (rank : 40) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1537 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9UPA5, O43161, Q7LGH3 | Gene names | BSN, KIAA0434, ZNF231 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein bassoon (Zinc finger protein 231). | |||||
|
ADIP_MOUSE
|
||||||
| NC score | 0.025436 (rank : 41) | θ value | 1.06291 (rank : 26) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 415 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VC66, Q8BG59, Q8C7X0, Q8K2F7 | Gene names | Ssx2ip | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Afadin- and alpha-actinin-binding protein (ADIP) (Afadin DIL domain- interacting protein). | |||||
|
FMNL_HUMAN
|
||||||
| NC score | 0.025082 (rank : 42) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | O95466, Q6DKG5, Q6IBP3, Q86UH1, Q8N671, Q8TDH1, Q96H10 | Gene names | FMNL1, C17orf1, C17orf1B, FMNL | |||
|
Domain Architecture |

|
|||||
| Description | Formin-like protein 1 (Leukocyte formin) (CLL-associated antigen KW- 13). | |||||
|
KISS1_HUMAN
|
||||||
| NC score | 0.023720 (rank : 43) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15726, Q9HBP1 | Gene names | KISS1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Metastasis-suppressor KiSS-1 precursor (Kisspeptin-1) [Contains: Metastin (Kisspeptin-54); Kisspeptin-14; Kisspeptin-13; Kisspeptin- 10]. | |||||
|
ATN1_HUMAN
|
||||||
| NC score | 0.023431 (rank : 44) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 676 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P54259, Q99495, Q99621, Q9UEK7 | Gene names | ATN1, DRPLA | |||
|
Domain Architecture |

|
|||||
| Description | Atrophin-1 (Dentatorubral-pallidoluysian atrophy protein). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.022597 (rank : 45) | θ value | 5.27518 (rank : 59) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
TMBI1_HUMAN
|
||||||
| NC score | 0.022180 (rank : 46) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q969X1, Q8N1R3, Q8TAM3, Q96K13 | Gene names | TMBIM1, RECS1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane BAX inhibitor motif-containing protein 1 (RECS1 protein homolog). | |||||
|
PRP2_MOUSE
|
||||||
| NC score | 0.021864 (rank : 47) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 347 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | P05142 | Gene names | Prh1, Prp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein MP-2 precursor. | |||||
|
CPSF6_HUMAN
|
||||||
| NC score | 0.021822 (rank : 48) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q16630, Q53ES1, Q9BSJ7, Q9BW18 | Gene names | CPSF6, CFIM68 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6 (Cleavage and polyadenylation specificity factor 68 kDa subunit) (CPSF 68 kDa subunit) (Pre-mRNA cleavage factor Im 68 kDa subunit) (Protein HPBRII- 4/7). | |||||
|
CO4A1_HUMAN
|
||||||
| NC score | 0.021594 (rank : 49) | θ value | 1.06291 (rank : 27) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P02462 | Gene names | COL4A1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(IV) chain precursor. | |||||
|
DAAM1_HUMAN
|
||||||
| NC score | 0.021202 (rank : 50) | θ value | 4.03905 (rank : 51) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 483 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9Y4D1, Q86U34, Q8N1Z8, Q8TB39 | Gene names | DAAM1, KIAA0666 | |||
|
Domain Architecture |

|
|||||
| Description | Disheveled-associated activator of morphogenesis 1. | |||||
|
CPSF6_MOUSE
|
||||||
| NC score | 0.021200 (rank : 51) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q6NVF9, Q8BX86, Q8BXI8 | Gene names | Cpsf6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cleavage and polyadenylation specificity factor 6. | |||||
|
EMID2_HUMAN
|
||||||
| NC score | 0.020965 (rank : 52) | θ value | 4.03905 (rank : 52) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 226 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q96A83 | Gene names | EMID2, COL26A1, EMU2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-1(XXVI) chain precursor (EMI domain-containing protein 2) (Protein Emu2) (Emilin and multimerin domain-containing protein 2). | |||||
|
NEUR4_HUMAN
|
||||||
| NC score | 0.020657 (rank : 53) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 33 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WWR8, Q96D64 | Gene names | NEU4 | |||
|
Domain Architecture |

|
|||||
| Description | Sialidase-4 (EC 3.2.1.18) (N-acetyl-alpha-neuraminidase 4). | |||||
|
SSBP3_HUMAN
|
||||||
| NC score | 0.020629 (rank : 54) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9BWW4, Q9BTM0, Q9BWW3 | Gene names | SSBP3, SSDP, SSDP1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein). | |||||
|
SSBP3_MOUSE
|
||||||
| NC score | 0.020629 (rank : 55) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9D032, Q99LC6, Q9EQP3 | Gene names | Ssbp3, Last, Ssdp1 | |||
|
Domain Architecture |

|
|||||
| Description | Single-stranded DNA-binding protein 3 (Sequence-specific single- stranded-DNA-binding protein) (Lck-associated signal transducer). | |||||
|
COIA1_MOUSE
|
||||||
| NC score | 0.019429 (rank : 56) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 481 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | P39061, Q60672, Q61437, Q62001, Q62002, Q9JK63 | Gene names | Col18a1 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-1(XVIII) chain precursor [Contains: Endostatin]. | |||||
|
PEF1_MOUSE
|
||||||
| NC score | 0.019348 (rank : 57) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 112 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8BFY6, Q8VCT5, Q9CYW8, Q9D934 | Gene names | Pef1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peflin (PEF protein with a long N-terminal hydrophobic domain) (Penta- EF hand domain-containing protein 1). | |||||
|
CO9A3_HUMAN
|
||||||
| NC score | 0.018782 (rank : 58) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q14050, Q13681, Q9H4G9, Q9UPE2 | Gene names | COL9A3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-3(IX) chain precursor. | |||||
|
CCD93_HUMAN
|
||||||
| NC score | 0.018513 (rank : 59) | θ value | 1.38821 (rank : 30) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 600 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q567U6, Q4LE78, Q53TJ2, Q8TBX5, Q9H6R5, Q9NV15 | Gene names | CCDC93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 93. | |||||
|
ANXA7_HUMAN
|
||||||
| NC score | 0.018229 (rank : 60) | θ value | 1.81305 (rank : 33) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 168 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P20073 | Gene names | ANXA7, ANX7, SNX | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A7 (Annexin VII) (Synexin). | |||||
|
HCN2_HUMAN
|
||||||
| NC score | 0.018070 (rank : 61) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UL51, O60742, O60743, O75267, Q9UBS2 | Gene names | HCN2, BCNG2 | |||
|
Domain Architecture |

|
|||||
| Description | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 (Brain cyclic nucleotide-gated channel 2) (BCNG-2). | |||||
|
CO4A3_HUMAN
|
||||||
| NC score | 0.017641 (rank : 62) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q01955, Q9BQT2 | Gene names | COL4A3 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-3(IV) chain precursor (Goodpasture antigen). | |||||
|
ANX11_MOUSE
|
||||||
| NC score | 0.016336 (rank : 63) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 200 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P97384 | Gene names | Anxa11, Anx11 | |||
|
Domain Architecture |

|
|||||
| Description | Annexin A11 (Annexin XI) (Calcyclin-associated annexin 50) (CAP-50). | |||||
|
CN140_MOUSE
|
||||||
| NC score | 0.015743 (rank : 64) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CCG1, Q8CCM8 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf140 homolog. | |||||
|
CCNK_MOUSE
|
||||||
| NC score | 0.014728 (rank : 65) | θ value | 5.27518 (rank : 58) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | O88874, Q8R068 | Gene names | Ccnk | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin-K. | |||||
|
CO9A2_MOUSE
|
||||||
| NC score | 0.014473 (rank : 66) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 279 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q07643 | Gene names | Col9a2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Collagen alpha-2(IX) chain precursor. | |||||
|
RERE_MOUSE
|
||||||
| NC score | 0.014407 (rank : 67) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 785 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q80TZ9 | Gene names | Rere, Atr2, Kiaa0458 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arginine-glutamic acid dipeptide repeats protein (Atrophin-2). | |||||
|
FOSB_HUMAN
|
||||||
| NC score | 0.013538 (rank : 68) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 141 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53539 | Gene names | FOSB, G0S3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein fosB (G0/G1 switch regulatory protein 3). | |||||
|
GSCR1_HUMAN
|
||||||
| NC score | 0.013067 (rank : 69) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9NZM4 | Gene names | GLTSCR1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Glioma tumor suppressor candidate region gene 1 protein. | |||||
|
GCNL2_MOUSE
|
||||||
| NC score | 0.012714 (rank : 70) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 146 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9JHD2 | Gene names | Gcn5l2 | |||
|
Domain Architecture |

|
|||||
| Description | General control of amino acid synthesis protein 5-like 2 (EC 2.3.1.48) (Histone acetyltransferase GCN5) (mmGCN5). | |||||
|
PRP1_HUMAN
|
||||||
| NC score | 0.012614 (rank : 71) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P04280, Q08805, Q15186, Q15187, Q15214, Q15215, Q16038 | Gene names | PRB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Basic salivary proline-rich protein 1 precursor (Salivary proline-rich protein) [Contains: Basic peptide IB-6; Peptide P-H]. | |||||
|
COBA2_HUMAN
|
||||||
| NC score | 0.012130 (rank : 72) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 408 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P13942, Q07751, Q13271, Q13272, Q13273, Q99866, Q9UIP9 | Gene names | COL11A2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
FOG1_HUMAN
|
||||||
| NC score | 0.011431 (rank : 73) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 981 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8IX07 | Gene names | ZFPM1, FOG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein ZFPM1 (Zinc finger protein multitype 1) (Friend of GATA protein 1) (Friend of GATA-1) (FOG-1). | |||||
|
IRX3_MOUSE
|
||||||
| NC score | 0.009139 (rank : 74) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 303 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P81067 | Gene names | Irx3, Irxb1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
IRX3_HUMAN
|
||||||
| NC score | 0.008972 (rank : 75) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P78415, Q7Z4A4, Q7Z4A5, Q8IVC6 | Gene names | IRX3, IRXB1 | |||
|
Domain Architecture |

|
|||||
| Description | Iroquois-class homeodomain protein IRX-3 (Iroquois homeobox protein 3) (Homeodomain protein IRXB1). | |||||
|
STRN3_MOUSE
|
||||||
| NC score | 0.008647 (rank : 76) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 309 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ERG2 | Gene names | Strn3, Gs2na, Sg2na | |||
|
Domain Architecture |

|
|||||
| Description | Striatin-3 (Cell-cycle autoantigen SG2NA) (S/G2 antigen). | |||||
|
RGS7_MOUSE
|
||||||
| NC score | 0.008508 (rank : 77) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O54829 | Gene names | Rgs7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
RGS7_HUMAN
|
||||||
| NC score | 0.008490 (rank : 78) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 82 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49802, Q8TD66, Q8TD67, Q9UNU7, Q9Y6B9 | Gene names | RGS7 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 7 (RGS7). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.006812 (rank : 79) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
LTBP2_MOUSE
|
||||||
| NC score | 0.005292 (rank : 80) | θ value | 4.03905 (rank : 53) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 617 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | O08999, Q8C6W9 | Gene names | Ltbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Latent-transforming growth factor beta-binding protein 2 precursor (LTBP-2). | |||||
|
LHX5_MOUSE
|
||||||
| NC score | 0.003716 (rank : 81) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 456 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P61375, P50459 | Gene names | Lhx5 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx5. | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.003683 (rank : 82) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
CADH4_HUMAN
|
||||||
| NC score | 0.002852 (rank : 83) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P55283, Q2M208, Q5VZ44, Q9BZ05 | Gene names | CDH4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
CADH4_MOUSE
|
||||||
| NC score | 0.002848 (rank : 84) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P39038 | Gene names | Cdh4 | |||
|
Domain Architecture |

|
|||||
| Description | Cadherin-4 precursor (Retinal-cadherin) (R-cadherin) (R-CAD). | |||||
|
TMPS6_HUMAN
|
||||||
| NC score | 0.001387 (rank : 85) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 85 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IU80, Q5TI06, Q6UXD8, Q8IUE2, Q8IXV8 | Gene names | TMPRSS6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||