Please be patient as the page loads
|
SEP10_HUMAN
|
||||||
| SwissProt Accessions | Q9P0V9, Q9HAH6 | Gene names | SEPT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-10. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SEP10_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 161 | |
| SwissProt Accessions | Q9P0V9, Q9HAH6 | Gene names | SEPT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-10. | |||||
|
SEP10_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.994087 (rank : 2) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q8C650, Q7TSA5 | Gene names | Sept10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-10. | |||||
|
SEP11_MOUSE
|
||||||
| θ value | 4.6918e-174 (rank : 3) | NC score | 0.968512 (rank : 7) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8C1B7, Q6P2K5, Q6P6I0 | Gene names | Sept11, D5Ertd606e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-11. | |||||
|
SEP11_HUMAN
|
||||||
| θ value | 8.84838e-173 (rank : 4) | NC score | 0.969359 (rank : 6) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NVA2, Q7L4N1 | Gene names | SEPT11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-11. | |||||
|
SEPT6_HUMAN
|
||||||
| θ value | 2.17943e-171 (rank : 5) | NC score | 0.980370 (rank : 3) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14141, Q5JTK0, Q969W5, Q96A13, Q96GR1, Q96P86, Q96P87 | Gene names | SEPT6, KIAA0128, SEP2 | |||
|
Domain Architecture |
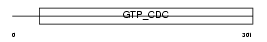
|
|||||
| Description | Septin-6. | |||||
|
SEPT6_MOUSE
|
||||||
| θ value | 4.11024e-170 (rank : 6) | NC score | 0.978646 (rank : 4) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9R1T4, Q91XH2, Q9CZ94 | Gene names | Sept6 | |||
|
Domain Architecture |
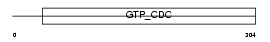
|
|||||
| Description | Septin-6. | |||||
|
SEPT8_HUMAN
|
||||||
| θ value | 5.93515e-169 (rank : 7) | NC score | 0.968060 (rank : 8) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q92599, Q8IX36, Q8IX37, Q9BVB3 | Gene names | SEPT8, KIAA0202 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-8. | |||||
|
SEPT8_MOUSE
|
||||||
| θ value | 4.25343e-167 (rank : 8) | NC score | 0.973393 (rank : 5) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8CHH9, Q80YC7, Q9ESF7 | Gene names | Sept8, Kiaa0202 | |||
|
Domain Architecture |
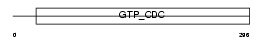
|
|||||
| Description | Septin-8. | |||||
|
SEPT7_HUMAN
|
||||||
| θ value | 6.73165e-80 (rank : 9) | NC score | 0.934331 (rank : 19) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q16181, Q6NX50 | Gene names | SEPT7, CDC10 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-7 (CDC10 protein homolog). | |||||
|
SEPT7_MOUSE
|
||||||
| θ value | 6.73165e-80 (rank : 10) | NC score | 0.933239 (rank : 20) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O55131 | Gene names | Sept7, Cdc10 | |||
|
Domain Architecture |
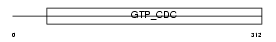
|
|||||
| Description | Septin-7 (CDC10 protein homolog). | |||||
|
SEPT3_HUMAN
|
||||||
| θ value | 1.71981e-67 (rank : 11) | NC score | 0.957619 (rank : 9) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UH03, Q6IBZ6, Q8N3P3 | Gene names | SEPT3, SEP3 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal-specific septin-3. | |||||
|
SEPT4_MOUSE
|
||||||
| θ value | 2.24614e-67 (rank : 12) | NC score | 0.946237 (rank : 17) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P28661, Q5ND15 | Gene names | Sept4, Pnutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-4 (Peanut-like protein 2) (Brain protein H5). | |||||
|
SEPT5_HUMAN
|
||||||
| θ value | 3.83135e-67 (rank : 13) | NC score | 0.949677 (rank : 14) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99719, O15251 | Gene names | SEPT5, PNUTL1 | |||
|
Domain Architecture |
|
|||||
| Description | Septin-5 (Peanut-like protein 1) (Cell division control-related protein 1) (CDCrel-1). | |||||
|
SEPT5_MOUSE
|
||||||
| θ value | 5.00389e-67 (rank : 14) | NC score | 0.950280 (rank : 13) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z2Q6, Q3UYG4, Q6PB74 | Gene names | Sept5, Pnutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-5 (Peanut-like protein 1) (Cell division control-related protein 1) (CDCrel-1). | |||||
|
SEPT4_HUMAN
|
||||||
| θ value | 8.53533e-67 (rank : 15) | NC score | 0.945508 (rank : 18) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43236, Q9UM58 | Gene names | SEPT4, PNUTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-4 (Peanut-like protein 2) (Brain protein H5) (Cell division control-related protein 2) (hCDCREL-2) (Bradeion beta) (CE5B3 beta) (Cerebral protein 7). | |||||
|
SEPT9_HUMAN
|
||||||
| θ value | 2.74572e-65 (rank : 16) | NC score | 0.866314 (rank : 22) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SEPT9_MOUSE
|
||||||
| θ value | 6.11685e-65 (rank : 17) | NC score | 0.923650 (rank : 21) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
SEPT1_HUMAN
|
||||||
| θ value | 3.03575e-64 (rank : 18) | NC score | 0.949332 (rank : 15) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WYJ6, Q8NEZ1, Q96EL4, Q9H285 | Gene names | SEPT1, PNUTL3 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-1 (LARP) (Serologically defined breast cancer antigen NY-BR- 24). | |||||
|
SEPT2_MOUSE
|
||||||
| θ value | 3.03575e-64 (rank : 19) | NC score | 0.950456 (rank : 12) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P42208, Q3U9Y5 | Gene names | Sept2, Nedd-5, Nedd5 | |||
|
Domain Architecture |
|
|||||
| Description | Septin-2 (Protein NEDD5) (Neural precursor cell expressed developmentally down-regulated protein 5). | |||||
|
SEPT2_HUMAN
|
||||||
| θ value | 3.96481e-64 (rank : 20) | NC score | 0.950935 (rank : 11) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15019, Q14132, Q8IUK9, Q96CB0 | Gene names | SEPT2, DIFF6, KIAA0158, NEDD5 | |||
|
Domain Architecture |
|
|||||
| Description | Septin-2 (Protein NEDD5). | |||||
|
SEPT3_MOUSE
|
||||||
| θ value | 5.7252e-63 (rank : 21) | NC score | 0.953259 (rank : 10) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z1S5 | Gene names | Sept3, Sep3 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal-specific septin-3. | |||||
|
SEPT1_MOUSE
|
||||||
| θ value | 1.19377e-60 (rank : 22) | NC score | 0.948750 (rank : 16) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P42209 | Gene names | Sept1, Diff6, Pnutl3 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-1 (Differentiation protein 6) (Diff6 protein). | |||||
|
GRAP1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 23) | NC score | 0.064627 (rank : 31) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
SWP70_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 24) | NC score | 0.084451 (rank : 23) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
GRAP1_HUMAN
|
||||||
| θ value | 0.0563607 (rank : 25) | NC score | 0.050967 (rank : 75) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
CTRO_MOUSE
|
||||||
| θ value | 0.0736092 (rank : 26) | NC score | 0.036302 (rank : 125) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 0.0961366 (rank : 27) | NC score | 0.061420 (rank : 39) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | 0.125558 (rank : 28) | NC score | 0.063057 (rank : 35) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | 0.163984 (rank : 29) | NC score | 0.080636 (rank : 26) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
CAPS2_HUMAN
|
||||||
| θ value | 0.279714 (rank : 30) | NC score | 0.029783 (rank : 137) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UW7, Q658Q2, Q7Z5T7, Q8IZW9, Q8N7M4, Q9H6P4, Q9HCI1, Q9NWK8 | Gene names | CADPS2, CAPS2, KIAA1591 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 2 (Calcium-dependent activator protein for secretion 2) (CAPS-2). | |||||
|
LMNB1_MOUSE
|
||||||
| θ value | 0.279714 (rank : 31) | NC score | 0.036592 (rank : 123) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P14733, Q61791 | Gene names | Lmnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
WBP11_HUMAN
|
||||||
| θ value | 0.279714 (rank : 32) | NC score | 0.080779 (rank : 25) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
WBP11_MOUSE
|
||||||
| θ value | 0.279714 (rank : 33) | NC score | 0.082068 (rank : 24) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.365318 (rank : 34) | NC score | 0.061225 (rank : 40) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
RRBP1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 35) | NC score | 0.056569 (rank : 53) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
LMNB1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 36) | NC score | 0.034502 (rank : 128) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P20700, Q3SYN7, Q96EI6 | Gene names | LMNB1, LMN2, LMNB | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 0.47712 (rank : 37) | NC score | 0.042807 (rank : 102) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
PKHG1_HUMAN
|
||||||
| θ value | 0.47712 (rank : 38) | NC score | 0.028218 (rank : 140) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULL1, Q5T1F2 | Gene names | PLEKHG1, KIAA1209 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family G member 1. | |||||
|
U2AFM_HUMAN
|
||||||
| θ value | 0.47712 (rank : 39) | NC score | 0.052864 (rank : 67) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
CAPS2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 40) | NC score | 0.028922 (rank : 139) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BYR5, O08903, Q66JM7, Q6PCL7, Q76I88, Q7TMM6, Q80ZV8, Q8BL25, Q8BY04, Q8K3K6 | Gene names | Cadps2, Caps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 2 (Calcium-dependent activator protein for secretion 2) (CAPS-2). | |||||
|
LRSM1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 41) | NC score | 0.034410 (rank : 129) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
MINK1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 42) | NC score | 0.007662 (rank : 178) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
MINK1_MOUSE
|
||||||
| θ value | 0.62314 (rank : 43) | NC score | 0.007681 (rank : 177) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
BEGIN_HUMAN
|
||||||
| θ value | 0.813845 (rank : 44) | NC score | 0.048546 (rank : 82) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9BUH8, Q9NPU3, Q9P282 | Gene names | BEGAIN, KIAA1446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
CP250_MOUSE
|
||||||
| θ value | 0.813845 (rank : 45) | NC score | 0.048563 (rank : 81) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
LIPA2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 46) | NC score | 0.048756 (rank : 79) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 0.813845 (rank : 47) | NC score | 0.045556 (rank : 91) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
SWP70_HUMAN
|
||||||
| θ value | 0.813845 (rank : 48) | NC score | 0.060146 (rank : 41) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
ZN291_HUMAN
|
||||||
| θ value | 0.813845 (rank : 49) | NC score | 0.045842 (rank : 89) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 50) | NC score | 0.058728 (rank : 44) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
INCE_HUMAN
|
||||||
| θ value | 1.06291 (rank : 51) | NC score | 0.048611 (rank : 80) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
KMHN1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 52) | NC score | 0.038468 (rank : 116) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
TACC3_MOUSE
|
||||||
| θ value | 1.06291 (rank : 53) | NC score | 0.042890 (rank : 101) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9JJ11, Q9WVK9 | Gene names | Tacc3, Aint | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ARNT-interacting protein). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 1.38821 (rank : 54) | NC score | 0.041530 (rank : 105) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CCD39_HUMAN
|
||||||
| θ value | 1.38821 (rank : 55) | NC score | 0.045702 (rank : 90) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
MEFV_HUMAN
|
||||||
| θ value | 1.38821 (rank : 56) | NC score | 0.011122 (rank : 169) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
OXR1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 57) | NC score | 0.017850 (rank : 157) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
PCNT_MOUSE
|
||||||
| θ value | 1.38821 (rank : 58) | NC score | 0.049590 (rank : 77) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
PLEC1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 59) | NC score | 0.033913 (rank : 131) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
RN168_HUMAN
|
||||||
| θ value | 1.38821 (rank : 60) | NC score | 0.019897 (rank : 154) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IYW5, Q8NA67, Q96NS4 | Gene names | RNF168 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 168. | |||||
|
TRHY_HUMAN
|
||||||
| θ value | 1.38821 (rank : 61) | NC score | 0.039934 (rank : 110) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
BICD1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 62) | NC score | 0.049035 (rank : 78) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
BICD1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 63) | NC score | 0.058319 (rank : 45) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
CAF1A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 64) | NC score | 0.038781 (rank : 114) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
CECR2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 65) | NC score | 0.013461 (rank : 164) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
FYB_MOUSE
|
||||||
| θ value | 1.81305 (rank : 66) | NC score | 0.018106 (rank : 156) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
GBP1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 67) | NC score | 0.024880 (rank : 144) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P32455 | Gene names | GBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 1 (GTP-binding protein 1) (Guanine nucleotide-binding protein 1) (GBP-1) (HuGBP-1). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 68) | NC score | 0.040464 (rank : 108) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
ITSN1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 69) | NC score | 0.041839 (rank : 104) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
MYO5A_MOUSE
|
||||||
| θ value | 1.81305 (rank : 70) | NC score | 0.027838 (rank : 141) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
RABE1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 71) | NC score | 0.046940 (rank : 86) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
SKIL_MOUSE
|
||||||
| θ value | 1.81305 (rank : 72) | NC score | 0.067677 (rank : 30) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q60665, Q60702, Q78E90, Q80VK5 | Gene names | Skil, Skir, Sno | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
TCRG1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 73) | NC score | 0.054092 (rank : 61) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
TCRG1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 74) | NC score | 0.056169 (rank : 54) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
TF2H1_HUMAN
|
||||||
| θ value | 1.81305 (rank : 75) | NC score | 0.025835 (rank : 143) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P32780, Q9H5K5, Q9NQD9 | Gene names | GTF2H1, BTF2 | |||
|
Domain Architecture |
|
|||||
| Description | TFIIH basal transcription factor complex p62 subunit (Basic transcription factor 62 kDa subunit) (BTF2-p62) (General transcription factor IIH polypeptide 1). | |||||
|
TF2H1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 76) | NC score | 0.025839 (rank : 142) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBA9, O35637 | Gene names | Gtf2h1 | |||
|
Domain Architecture |
|
|||||
| Description | TFIIH basal transcription factor complex p62 subunit (Basic transcription factor 62 kDa subunit) (BTF2-p62) (General transcription factor IIH polypeptide 1). | |||||
|
BEGIN_MOUSE
|
||||||
| θ value | 2.36792 (rank : 77) | NC score | 0.041438 (rank : 106) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q68EF6 | Gene names | Begain | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
IF3A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 78) | NC score | 0.040175 (rank : 109) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 79) | NC score | 0.023399 (rank : 148) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 80) | NC score | 0.046497 (rank : 88) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 81) | NC score | 0.044493 (rank : 97) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
TRI32_MOUSE
|
||||||
| θ value | 2.36792 (rank : 82) | NC score | 0.010653 (rank : 174) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CH72, Q8K055 | Gene names | Trim32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-). | |||||
|
U2AFM_MOUSE
|
||||||
| θ value | 2.36792 (rank : 83) | NC score | 0.045025 (rank : 93) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
BICD2_HUMAN
|
||||||
| θ value | 3.0926 (rank : 84) | NC score | 0.053706 (rank : 63) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
BICD2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 85) | NC score | 0.057898 (rank : 47) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
GBP2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 86) | NC score | 0.029043 (rank : 138) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Z0E6, Q8CIC6, Q921N2, Q9R1I0 | Gene names | Gbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 2 (GTP-binding protein 2) (Guanine nucleotide-binding protein 2) (GBP-2) (mGBP2) (mGBP-2). | |||||
|
HIP1R_MOUSE
|
||||||
| θ value | 3.0926 (rank : 87) | NC score | 0.045420 (rank : 92) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
IF2P_HUMAN
|
||||||
| θ value | 3.0926 (rank : 88) | NC score | 0.034369 (rank : 130) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
MICA1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 89) | NC score | 0.021759 (rank : 151) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
PININ_MOUSE
|
||||||
| θ value | 3.0926 (rank : 90) | NC score | 0.035446 (rank : 127) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
RAB15_HUMAN
|
||||||
| θ value | 3.0926 (rank : 91) | NC score | 0.013916 (rank : 163) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P59190, Q86TX7, Q8IW89 | Gene names | RAB15 | |||
|
Domain Architecture |
|
|||||
| Description | Ras-related protein Rab-15. | |||||
|
RNF8_MOUSE
|
||||||
| θ value | 3.0926 (rank : 92) | NC score | 0.024161 (rank : 146) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8VC56, Q9JK13 | Gene names | Rnf8 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase protein RNF8 (EC 6.3.2.-) (RING finger protein 8) (AIP37) (ActA-interacting protein 37) (LaXp180). | |||||
|
SM1L2_MOUSE
|
||||||
| θ value | 3.0926 (rank : 93) | NC score | 0.033629 (rank : 132) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
U2AFL_HUMAN
|
||||||
| θ value | 3.0926 (rank : 94) | NC score | 0.044681 (rank : 94) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
CING_HUMAN
|
||||||
| θ value | 4.03905 (rank : 95) | NC score | 0.052532 (rank : 69) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
DBP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 96) | NC score | 0.011745 (rank : 168) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60925, Q8VCX3 | Gene names | Dbp | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 4.03905 (rank : 97) | NC score | 0.055654 (rank : 56) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
DMD_HUMAN
|
||||||
| θ value | 4.03905 (rank : 98) | NC score | 0.041368 (rank : 107) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
LIPA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 99) | NC score | 0.037912 (rank : 119) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
LIPA3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 100) | NC score | 0.037673 (rank : 121) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P60469 | Gene names | Ppfia3 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
MAT1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 101) | NC score | 0.021775 (rank : 150) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P51949, Q3TZP0, Q9D8A0, Q9D8D2 | Gene names | Mnat1, Mat1 | |||
|
Domain Architecture |
|
|||||
| Description | CDK-activating kinase assembly factor MAT1 (RING finger protein MAT1) (Menage a trois) (CDK7/cyclin H assembly factor) (p36) (p35). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 102) | NC score | 0.044079 (rank : 99) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
RAB10_HUMAN
|
||||||
| θ value | 4.03905 (rank : 103) | NC score | 0.010765 (rank : 171) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P61026, O88386, Q6IA52, Q9D7X6, Q9H0T3 | Gene names | RAB10 | |||
|
Domain Architecture |
|
|||||
| Description | Ras-related protein Rab-10. | |||||
|
RAB10_MOUSE
|
||||||
| θ value | 4.03905 (rank : 104) | NC score | 0.010765 (rank : 172) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P61027, O88386, Q9D7X6 | Gene names | Rab10 | |||
|
Domain Architecture |
|
|||||
| Description | Ras-related protein Rab-10. | |||||
|
VP13A_HUMAN
|
||||||
| θ value | 4.03905 (rank : 105) | NC score | 0.036158 (rank : 126) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96RL7, Q5JSX9, Q5JSY0, Q5VYR5, Q702P4, Q709D0, Q86YF8, Q96S61, Q9H995, Q9Y2J1 | Gene names | VPS13A, CHAC, KIAA0986 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 13A (Chorein) (Chorea- acanthocytosis protein). | |||||
|
BRE1A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 106) | NC score | 0.031878 (rank : 135) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
CCD91_HUMAN
|
||||||
| θ value | 5.27518 (rank : 107) | NC score | 0.063359 (rank : 33) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
CD2L1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 108) | NC score | 0.002846 (rank : 185) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 5.27518 (rank : 109) | NC score | 0.063153 (rank : 34) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CTGE5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 110) | NC score | 0.044462 (rank : 98) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
DBP_HUMAN
|
||||||
| θ value | 5.27518 (rank : 111) | NC score | 0.010936 (rank : 170) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q10586 | Gene names | DBP | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein) (TAXREB302). | |||||
|
FYB_HUMAN
|
||||||
| θ value | 5.27518 (rank : 112) | NC score | 0.013372 (rank : 165) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 113) | NC score | 0.044615 (rank : 96) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
GCP60_HUMAN
|
||||||
| θ value | 5.27518 (rank : 114) | NC score | 0.031685 (rank : 136) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9H3P7, Q5VTJ0, Q6P9F1, Q8IZC5, Q8N4D6, Q9H6U3 | Gene names | ACBD3, GCP60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | 5.27518 (rank : 115) | NC score | 0.046587 (rank : 87) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
MAT1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 116) | NC score | 0.020631 (rank : 152) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P51948, Q15817 | Gene names | MNAT1, CAP35, MAT1, RNF66 | |||
|
Domain Architecture |
|
|||||
| Description | CDK-activating kinase assembly factor MAT1 (RING finger protein MAT1) (Menage a trois) (CDK7/cyclin H assembly factor) (p36) (p35) (Cyclin G1-interacting protein) (RING finger protein 66). | |||||
|
MYO5B_HUMAN
|
||||||
| θ value | 5.27518 (rank : 117) | NC score | 0.022677 (rank : 149) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
NIN_MOUSE
|
||||||
| θ value | 5.27518 (rank : 118) | NC score | 0.039446 (rank : 111) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
NUCB2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 119) | NC score | 0.038394 (rank : 117) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 5.27518 (rank : 120) | NC score | 0.044633 (rank : 95) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
UBXD8_HUMAN
|
||||||
| θ value | 5.27518 (rank : 121) | NC score | 0.016622 (rank : 158) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96CS3, O94963, Q8IUF2, Q9BRP2, Q9BVM7 | Gene names | UBXD8, ETEA, KIAA0887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 8 (Protein ETEA). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | 6.88961 (rank : 122) | NC score | 0.047266 (rank : 85) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 116 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
BRD3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 123) | NC score | 0.010264 (rank : 175) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
CN145_MOUSE
|
||||||
| θ value | 6.88961 (rank : 124) | NC score | 0.038972 (rank : 113) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
CP135_MOUSE
|
||||||
| θ value | 6.88961 (rank : 125) | NC score | 0.043364 (rank : 100) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
ITSN2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 126) | NC score | 0.038012 (rank : 118) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 127) | NC score | 0.052129 (rank : 72) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
NAL13_HUMAN
|
||||||
| θ value | 6.88961 (rank : 128) | NC score | 0.002554 (rank : 186) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
OFD1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 129) | NC score | 0.062032 (rank : 37) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
PP16A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 130) | NC score | -0.000514 (rank : 188) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q923M0, Q4V9Z1 | Gene names | Ppp1r16a, Mypt3 | |||
|
Domain Architecture |
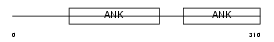
|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase targeting subunit 3). | |||||
|
RAB15_MOUSE
|
||||||
| θ value | 6.88961 (rank : 131) | NC score | 0.010680 (rank : 173) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K386 | Gene names | Rab15 | |||
|
Domain Architecture |
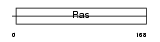
|
|||||
| Description | Ras-related protein Rab-15. | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 6.88961 (rank : 132) | NC score | 0.038739 (rank : 115) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
REST_HUMAN
|
||||||
| θ value | 6.88961 (rank : 133) | NC score | 0.047428 (rank : 84) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
RPGR1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 134) | NC score | 0.023536 (rank : 147) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
TWST2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 135) | NC score | 0.007281 (rank : 180) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 136) | NC score | 0.007296 (rank : 179) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
U2AFL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 137) | NC score | 0.039071 (rank : 112) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
UBXD8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 138) | NC score | 0.016076 (rank : 159) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3TDN2, Q3U9W2, Q80TP7, Q80W42 | Gene names | Ubxd8, Kiaa0887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 8. | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 8.99809 (rank : 139) | NC score | 0.058170 (rank : 46) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CD2L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 140) | NC score | 0.002507 (rank : 187) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 141) | NC score | 0.036489 (rank : 124) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 142) | NC score | 0.037834 (rank : 120) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
GCP60_MOUSE
|
||||||
| θ value | 8.99809 (rank : 143) | NC score | 0.033164 (rank : 133) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8BMP6, O35371 | Gene names | Acbd3, Gcp60, Pap7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
GDIT_HUMAN
|
||||||
| θ value | 8.99809 (rank : 144) | NC score | 0.004579 (rank : 182) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99819, Q4TT69, Q96S29 | Gene names | ARHGDIG | |||
|
Domain Architecture |
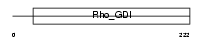
|
|||||
| Description | Rho GDP-dissociation inhibitor 3 (Rho GDI 3) (Rho-GDI gamma). | |||||
|
GOGA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 145) | NC score | 0.037642 (rank : 122) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 146) | NC score | 0.064289 (rank : 32) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
INCE_MOUSE
|
||||||
| θ value | 8.99809 (rank : 147) | NC score | 0.042322 (rank : 103) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
KIF5C_MOUSE
|
||||||
| θ value | 8.99809 (rank : 148) | NC score | 0.032838 (rank : 134) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
MOES_HUMAN
|
||||||
| θ value | 8.99809 (rank : 149) | NC score | 0.020462 (rank : 153) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
MYO6_HUMAN
|
||||||
| θ value | 8.99809 (rank : 150) | NC score | 0.018593 (rank : 155) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
PTN2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 151) | NC score | 0.006322 (rank : 181) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q06180, Q3V259, Q922E7 | Gene names | Ptpn2, Ptpt | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-2) (MPTP). | |||||
|
RAB13_HUMAN
|
||||||
| θ value | 8.99809 (rank : 152) | NC score | 0.008146 (rank : 176) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51153, Q96GU4 | Gene names | RAB13 | |||
|
Domain Architecture |
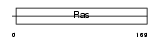
|
|||||
| Description | Ras-related protein Rab-13. | |||||
|
RAB1A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 153) | NC score | 0.012891 (rank : 166) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P62820, P11476, Q96N61, Q9Y3T2 | Gene names | RAB1A, RAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-related protein Rab-1A (YPT1-related protein). | |||||
|
RAB1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 154) | NC score | 0.012891 (rank : 167) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P62821, P11476, Q3TX44, Q811M4, Q96N61, Q9Y3T2 | Gene names | Rab1A, Rab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-related protein Rab-1A (YPT1-related protein). | |||||
|
RAB1B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 155) | NC score | 0.014530 (rank : 162) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H0U4 | Gene names | RAB1B | |||
|
Domain Architecture |
|
|||||
| Description | Ras-related protein Rab-1B. | |||||
|
RAB1B_MOUSE
|
||||||
| θ value | 8.99809 (rank : 156) | NC score | 0.014547 (rank : 161) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D1G1, Q3U0N1 | Gene names | Rab1b | |||
|
Domain Architecture |
|
|||||
| Description | Ras-related protein Rab-1B. | |||||
|
RAD50_MOUSE
|
||||||
| θ value | 8.99809 (rank : 157) | NC score | 0.048278 (rank : 83) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
REN3A_HUMAN
|
||||||
| θ value | 8.99809 (rank : 158) | NC score | 0.024840 (rank : 145) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H1J1, Q5T8C3, Q5T8C9, Q7Z6N3, Q86YK1, Q9BZI8 | Gene names | UPF3A, RENT3A, UPF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3A (Nonsense mRNA reducing factor 3A) (Up-frameshift suppressor 3 homolog A) (hUpf3). | |||||
|
SUV91_MOUSE
|
||||||
| θ value | 8.99809 (rank : 159) | NC score | 0.004491 (rank : 183) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
TAF3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 160) | NC score | 0.015121 (rank : 160) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
UBP37_HUMAN
|
||||||
| θ value | 8.99809 (rank : 161) | NC score | 0.003453 (rank : 184) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
APOA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.059361 (rank : 42) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P02647, Q6LDN9, Q6Q785 | Gene names | APOA1 | |||
|
Domain Architecture |
|
|||||
| Description | Apolipoprotein A-I precursor (Apo-AI) (ApoA-I) [Contains: Apolipoprotein A-I(1-242)]. | |||||
|
CCD41_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.059127 (rank : 43) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CCD46_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.056870 (rank : 51) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CCD96_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.055057 (rank : 58) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
CE290_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.057890 (rank : 48) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CEP63_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.051041 (rank : 74) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CI039_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.052934 (rank : 66) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.062624 (rank : 36) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CP135_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.052387 (rank : 70) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.053147 (rank : 64) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.050012 (rank : 76) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
HIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.052010 (rank : 73) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
MAP1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.055384 (rank : 57) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
NAF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.055731 (rank : 55) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
NAF1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.053131 (rank : 65) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
NUF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.054726 (rank : 59) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
PHLB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.057262 (rank : 49) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
PHLB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.056997 (rank : 50) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
RUFY2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.056589 (rank : 52) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.054368 (rank : 60) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.053736 (rank : 62) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.069013 (rank : 29) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
SMC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.061658 (rank : 38) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
SPC25_HUMAN
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.076611 (rank : 27) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HBM1 | Gene names | SPBC25, SPC25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc25 (hSpc25). | |||||
|
STMN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.052814 (rank : 68) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
STMN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.052356 (rank : 71) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
ZWINT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.071605 (rank : 28) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9CQU5, Q3UYT8, Q91VI4, Q9D0W9 | Gene names | Zwint, D10Ertd749e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ZW10 interactor (ZW10-interacting protein 1) (Zwint-1). | |||||
|
SEP10_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 161 | Shared Neighborhood Hits | 161 | |
| SwissProt Accessions | Q9P0V9, Q9HAH6 | Gene names | SEPT10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-10. | |||||
|
SEP10_MOUSE
|
||||||
| NC score | 0.994087 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 145 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q8C650, Q7TSA5 | Gene names | Sept10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-10. | |||||
|
SEPT6_HUMAN
|
||||||
| NC score | 0.980370 (rank : 3) | θ value | 2.17943e-171 (rank : 5) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 220 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14141, Q5JTK0, Q969W5, Q96A13, Q96GR1, Q96P86, Q96P87 | Gene names | SEPT6, KIAA0128, SEP2 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-6. | |||||
|
SEPT6_MOUSE
|
||||||
| NC score | 0.978646 (rank : 4) | θ value | 4.11024e-170 (rank : 6) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 244 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q9R1T4, Q91XH2, Q9CZ94 | Gene names | Sept6 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-6. | |||||
|
SEPT8_MOUSE
|
||||||
| NC score | 0.973393 (rank : 5) | θ value | 4.25343e-167 (rank : 8) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 66 | |
| SwissProt Accessions | Q8CHH9, Q80YC7, Q9ESF7 | Gene names | Sept8, Kiaa0202 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-8. | |||||
|
SEP11_HUMAN
|
||||||
| NC score | 0.969359 (rank : 6) | θ value | 8.84838e-173 (rank : 4) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9NVA2, Q7L4N1 | Gene names | SEPT11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-11. | |||||
|
SEP11_MOUSE
|
||||||
| NC score | 0.968512 (rank : 7) | θ value | 4.6918e-174 (rank : 3) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 300 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q8C1B7, Q6P2K5, Q6P6I0 | Gene names | Sept11, D5Ertd606e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-11. | |||||
|
SEPT8_HUMAN
|
||||||
| NC score | 0.968060 (rank : 8) | θ value | 5.93515e-169 (rank : 7) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 70 | |
| SwissProt Accessions | Q92599, Q8IX36, Q8IX37, Q9BVB3 | Gene names | SEPT8, KIAA0202 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-8. | |||||
|
SEPT3_HUMAN
|
||||||
| NC score | 0.957619 (rank : 9) | θ value | 1.71981e-67 (rank : 11) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9UH03, Q6IBZ6, Q8N3P3 | Gene names | SEPT3, SEP3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal-specific septin-3. | |||||
|
SEPT3_MOUSE
|
||||||
| NC score | 0.953259 (rank : 10) | θ value | 5.7252e-63 (rank : 21) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | Q9Z1S5 | Gene names | Sept3, Sep3 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal-specific septin-3. | |||||
|
SEPT2_HUMAN
|
||||||
| NC score | 0.950935 (rank : 11) | θ value | 3.96481e-64 (rank : 20) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q15019, Q14132, Q8IUK9, Q96CB0 | Gene names | SEPT2, DIFF6, KIAA0158, NEDD5 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-2 (Protein NEDD5). | |||||
|
SEPT2_MOUSE
|
||||||
| NC score | 0.950456 (rank : 12) | θ value | 3.03575e-64 (rank : 19) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 105 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P42208, Q3U9Y5 | Gene names | Sept2, Nedd-5, Nedd5 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-2 (Protein NEDD5) (Neural precursor cell expressed developmentally down-regulated protein 5). | |||||
|
SEPT5_MOUSE
|
||||||
| NC score | 0.950280 (rank : 13) | θ value | 5.00389e-67 (rank : 14) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Z2Q6, Q3UYG4, Q6PB74 | Gene names | Sept5, Pnutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-5 (Peanut-like protein 1) (Cell division control-related protein 1) (CDCrel-1). | |||||
|
SEPT5_HUMAN
|
||||||
| NC score | 0.949677 (rank : 14) | θ value | 3.83135e-67 (rank : 13) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q99719, O15251 | Gene names | SEPT5, PNUTL1 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-5 (Peanut-like protein 1) (Cell division control-related protein 1) (CDCrel-1). | |||||
|
SEPT1_HUMAN
|
||||||
| NC score | 0.949332 (rank : 15) | θ value | 3.03575e-64 (rank : 18) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 65 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q8WYJ6, Q8NEZ1, Q96EL4, Q9H285 | Gene names | SEPT1, PNUTL3 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-1 (LARP) (Serologically defined breast cancer antigen NY-BR- 24). | |||||
|
SEPT1_MOUSE
|
||||||
| NC score | 0.948750 (rank : 16) | θ value | 1.19377e-60 (rank : 22) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P42209 | Gene names | Sept1, Diff6, Pnutl3 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-1 (Differentiation protein 6) (Diff6 protein). | |||||
|
SEPT4_MOUSE
|
||||||
| NC score | 0.946237 (rank : 17) | θ value | 2.24614e-67 (rank : 12) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | P28661, Q5ND15 | Gene names | Sept4, Pnutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-4 (Peanut-like protein 2) (Brain protein H5). | |||||
|
SEPT4_HUMAN
|
||||||
| NC score | 0.945508 (rank : 18) | θ value | 8.53533e-67 (rank : 15) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 22 | |
| SwissProt Accessions | O43236, Q9UM58 | Gene names | SEPT4, PNUTL2 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-4 (Peanut-like protein 2) (Brain protein H5) (Cell division control-related protein 2) (hCDCREL-2) (Bradeion beta) (CE5B3 beta) (Cerebral protein 7). | |||||
|
SEPT7_HUMAN
|
||||||
| NC score | 0.934331 (rank : 19) | θ value | 6.73165e-80 (rank : 9) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 100 | |
| SwissProt Accessions | Q16181, Q6NX50 | Gene names | SEPT7, CDC10 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-7 (CDC10 protein homolog). | |||||
|
SEPT7_MOUSE
|
||||||
| NC score | 0.933239 (rank : 20) | θ value | 6.73165e-80 (rank : 10) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 327 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | O55131 | Gene names | Sept7, Cdc10 | |||
|
Domain Architecture |

|
|||||
| Description | Septin-7 (CDC10 protein homolog). | |||||
|
SEPT9_MOUSE
|
||||||
| NC score | 0.923650 (rank : 21) | θ value | 6.11685e-65 (rank : 17) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q80UG5, Q3URP2, Q80TM7, Q9QYX9 | Gene names | Sept9, Kiaa0991, Sint1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (SL3-3 integration site 1 protein). | |||||
|
SEPT9_HUMAN
|
||||||
| NC score | 0.866314 (rank : 22) | θ value | 2.74572e-65 (rank : 16) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q9UHD8, Q96QF3, Q96QF4, Q96QF5, Q9HA04, Q9UG40, Q9Y5W4 | Gene names | SEPT9, KIAA0991, MSF | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Septin-9 (MLL septin-like fusion protein) (MLL septin-like fusion protein MSF-A) (Ovarian/Breast septin) (Ov/Br septin) (Septin D1). | |||||
|
SWP70_MOUSE
|
||||||
| NC score | 0.084451 (rank : 23) | θ value | 0.0330416 (rank : 24) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1097 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q6A028, O88443, Q3TQR6, Q3UCA3, Q6P1D0 | Gene names | Swap70, Kiaa0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
WBP11_MOUSE
|
||||||
| NC score | 0.082068 (rank : 24) | θ value | 0.279714 (rank : 33) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q923D5, O88539, Q3UMD8, Q8VDI0 | Gene names | Wbp11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11). | |||||
|
WBP11_HUMAN
|
||||||
| NC score | 0.080779 (rank : 25) | θ value | 0.279714 (rank : 32) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q9Y2W2, Q96AY8 | Gene names | WBP11, NPWBP, SNP70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | WW domain-binding protein 11 (WBP-11) (SH3 domain-binding protein SNP70) (Npw38-binding protein) (NpwBP). | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.080636 (rank : 26) | θ value | 0.163984 (rank : 29) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
SPC25_HUMAN
|
||||||
| NC score | 0.076611 (rank : 27) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9HBM1 | Gene names | SPBC25, SPC25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc25 (hSpc25). | |||||
|
ZWINT_MOUSE
|
||||||
| NC score | 0.071605 (rank : 28) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 497 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | Q9CQU5, Q3UYT8, Q91VI4, Q9D0W9 | Gene names | Zwint, D10Ertd749e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ZW10 interactor (ZW10-interacting protein 1) (Zwint-1). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.069013 (rank : 29) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
SKIL_MOUSE
|
||||||
| NC score | 0.067677 (rank : 30) | θ value | 1.81305 (rank : 72) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | Q60665, Q60702, Q78E90, Q80VK5 | Gene names | Skil, Skir, Sno | |||
|
Domain Architecture |

|
|||||
| Description | Ski-like protein (Ski-related protein) (Ski-related oncogene). | |||||
|
GRAP1_MOUSE
|
||||||
| NC score | 0.064627 (rank : 31) | θ value | 0.0330416 (rank : 23) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.064289 (rank : 32) | θ value | 8.99809 (rank : 146) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 122 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
CCD91_HUMAN
|
||||||
| NC score | 0.063359 (rank : 33) | θ value | 5.27518 (rank : 107) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q7Z6B0, Q68D43, Q6IA78, Q8NEN7, Q9NUW9 | Gene names | CCDC91, GGABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 91 (GGA-binding partner) (p56 accessory protein). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.063153 (rank : 34) | θ value | 5.27518 (rank : 109) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.063057 (rank : 35) | θ value | 0.125558 (rank : 28) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.062624 (rank : 36) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 112 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
OFD1_HUMAN
|
||||||
| NC score | 0.062032 (rank : 37) | θ value | 6.88961 (rank : 129) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 87 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.061658 (rank : 38) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.061420 (rank : 39) | θ value | 0.0961366 (rank : 27) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.061225 (rank : 40) | θ value | 0.365318 (rank : 34) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
SWP70_HUMAN
|
||||||
| NC score | 0.060146 (rank : 41) | θ value | 0.813845 (rank : 48) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 973 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9UH65, O75135, Q7LCY6, Q9P061, Q9P0Z8 | Gene names | SWAP70, KIAA0640 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Switch-associated protein 70 (SWAP-70). | |||||
|
APOA1_HUMAN
|
||||||
| NC score | 0.059361 (rank : 42) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | P02647, Q6LDN9, Q6Q785 | Gene names | APOA1 | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein A-I precursor (Apo-AI) (ApoA-I) [Contains: Apolipoprotein A-I(1-242)]. | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.059127 (rank : 43) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.058728 (rank : 44) | θ value | 1.06291 (rank : 50) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 118 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
BICD1_MOUSE
|
||||||
| NC score | 0.058319 (rank : 45) | θ value | 1.81305 (rank : 63) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.058170 (rank : 46) | θ value | 8.99809 (rank : 139) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
BICD2_MOUSE
|
||||||
| NC score | 0.057898 (rank : 47) | θ value | 3.0926 (rank : 85) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.057890 (rank : 48) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
PHLB2_HUMAN
|
||||||
| NC score | 0.057262 (rank : 49) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q86SQ0, Q59EA8, Q68CY3, Q6NT98, Q8N8U8, Q8NAB1, Q8NCU5 | Gene names | PHLDB2, LL5B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
PHLB2_MOUSE
|
||||||
| NC score | 0.056997 (rank : 50) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 99 | |
| SwissProt Accessions | Q8K1N2, Q3TNI3, Q80Y16, Q8BKV3 | Gene names | Phldb2, Ll5b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology-like domain family B member 2 (Protein LL5-beta). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.056870 (rank : 51) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
RUFY2_HUMAN
|
||||||
| NC score | 0.056589 (rank : 52) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1208 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q8WXA3, Q96P51, Q9P1Z1 | Gene names | RUFY2, KIAA1537, RABIP4R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RUN and FYVE domain-containing protein 2 (Rab4-interacting protein related). | |||||
|
RRBP1_MOUSE
|
||||||
| NC score | 0.056569 (rank : 53) | θ value | 0.365318 (rank : 35) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1650 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q99PL5, Q99PK5, Q99PK6, Q99PK7, Q99PK8, Q99PK9, Q99PL0, Q99PL1, Q99PL2, Q99PL3, Q99PL4, Q9CS20 | Gene names | Rrbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Ribosome-binding protein 1 (Ribosome receptor protein) (mRRp). | |||||
|
TCRG1_MOUSE
|
||||||
| NC score | 0.056169 (rank : 54) | θ value | 1.81305 (rank : 74) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 947 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q8CGF7, Q61051, Q8C490, Q8CHT8, Q9R0R5 | Gene names | Tcerg1, Taf2s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150) (p144) (Formin- binding protein 28) (FBP 28). | |||||
|
NAF1_HUMAN
|
||||||
| NC score | 0.055731 (rank : 55) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q15025, O76008, Q96EL9, Q99833, Q9H1J3 | Gene names | TNIP1, KIAA0113, NAF1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (HIV-1 Nef-interacting protein) (Virion-associated nuclear shuttling protein) (VAN) (hVAN) (Nip40-1). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.055654 (rank : 56) | θ value | 4.03905 (rank : 97) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
MAP1A_HUMAN
|
||||||
| NC score | 0.055384 (rank : 57) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1626 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | P78559, O95643, Q12973, Q15882, Q9UJT4 | Gene names | MAP1A | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 1A (MAP 1A) (Proliferation-related protein p80) [Contains: MAP1 light chain LC2]. | |||||
|
CCD96_MOUSE
|
||||||
| NC score | 0.055057 (rank : 58) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 704 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9CR92 | Gene names | Ccdc96 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 96. | |||||
|
NUF2_HUMAN
|
||||||
| NC score | 0.054726 (rank : 59) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.054368 (rank : 60) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
TCRG1_HUMAN
|
||||||
| NC score | 0.054092 (rank : 61) | θ value | 1.81305 (rank : 73) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 929 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | O14776 | Gene names | TCERG1, CA150, TAF2S | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription elongation regulator 1 (TATA box-binding protein- associated factor 2S) (Transcription factor CA150). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.053736 (rank : 62) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
BICD2_HUMAN
|
||||||
| NC score | 0.053706 (rank : 63) | θ value | 3.0926 (rank : 84) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.053147 (rank : 64) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
NAF1_MOUSE
|
||||||
| NC score | 0.053131 (rank : 65) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 852 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q9WUU8, Q922A9, Q922F7, Q9EPP8, Q9R0X3 | Gene names | Tnip1, Abin, Naf1 | |||
|
Domain Architecture |

|
|||||
| Description | Nef-associated factor 1 (Naf1) (TNFAIP3-interacting protein 1) (A20- binding inhibitor of NF-kappa-B activation) (ABIN) (Virion-associated nuclear shuttling protein) (VAN) (mVAN). | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.052934 (rank : 66) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
U2AFM_HUMAN
|
||||||
| NC score | 0.052864 (rank : 67) | θ value | 0.47712 (rank : 39) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 577 | Shared Neighborhood Hits | 76 | |
| SwissProt Accessions | Q15696 | Gene names | ZRSR2, U2AF1-RS2, U2AF1L2, U2AF1RS2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2) (NY-REN-20 antigen). | |||||
|
STMN1_HUMAN
|
||||||
| NC score | 0.052814 (rank : 68) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 554 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | P16949 | Gene names | STMN1, LAP18, OP18 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Protein Pr22). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.052532 (rank : 69) | θ value | 4.03905 (rank : 95) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.052387 (rank : 70) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
STMN1_MOUSE
|
||||||
| NC score | 0.052356 (rank : 71) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 560 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | P54227 | Gene names | Stmn1, Lag, Lap18, Pr22 | |||
|
Domain Architecture |

|
|||||
| Description | Stathmin (Phosphoprotein p19) (pp19) (Oncoprotein 18) (Op18) (Leukemia-associated phosphoprotein p18) (pp17) (Prosolin) (Metablastin) (Pr22 protein) (Leukemia-associated gene protein). | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.052129 (rank : 72) | θ value | 6.88961 (rank : 127) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
HIP1_HUMAN
|
||||||
| NC score | 0.052010 (rank : 73) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1016 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | O00291, O00328, Q8TDL4 | Gene names | HIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1 (HIP-I). | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.051041 (rank : 74) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 102 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
GRAP1_HUMAN
|
||||||
| NC score | 0.050967 (rank : 75) | θ value | 0.0563607 (rank : 25) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.050012 (rank : 76) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
PCNT_MOUSE
|
||||||
| NC score | 0.049590 (rank : 77) | θ value | 1.38821 (rank : 58) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1207 | Shared Neighborhood Hits | 105 | |
| SwissProt Accessions | P48725 | Gene names | Pcnt, Pcnt2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pericentrin. | |||||
|
BICD1_HUMAN
|
||||||
| NC score | 0.049035 (rank : 78) | θ value | 1.81305 (rank : 62) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
LIPA2_HUMAN
|
||||||
| NC score | 0.048756 (rank : 79) | θ value | 0.813845 (rank : 46) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
INCE_HUMAN
|
||||||
| NC score | 0.048611 (rank : 80) | θ value | 1.06291 (rank : 51) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 978 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9NQS7, Q5Y192 | Gene names | INCENP | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.048563 (rank : 81) | θ value | 0.813845 (rank : 45) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 113 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
BEGIN_HUMAN
|
||||||
| NC score | 0.048546 (rank : 82) | θ value | 0.813845 (rank : 44) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 454 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q9BUH8, Q9NPU3, Q9P282 | Gene names | BEGAIN, KIAA1446 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.048278 (rank : 83) | θ value | 8.99809 (rank : 157) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.047428 (rank : 84) | θ value | 6.88961 (rank : 133) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.047266 (rank : 85) | θ value | 6.88961 (rank : 122) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 116 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
RABE1_MOUSE
|
||||||
| NC score | 0.046940 (rank : 86) | θ value | 1.81305 (rank : 71) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.046587 (rank : 87) | θ value | 5.27518 (rank : 115) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.046497 (rank : 88) | θ value | 2.36792 (rank : 80) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
ZN291_HUMAN
|
||||||
| NC score | 0.045842 (rank : 89) | θ value | 0.813845 (rank : 49) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 78 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.045702 (rank : 90) | θ value | 1.38821 (rank : 55) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 96 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.045556 (rank : 91) | θ value | 0.813845 (rank : 47) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
HIP1R_MOUSE
|
||||||
| NC score | 0.045420 (rank : 92) | θ value | 3.0926 (rank : 87) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 88 | |
| SwissProt Accessions | Q9JKY5 | Gene names | Hip1r | |||
|
Domain Architecture |

|
|||||
| Description | Huntingtin-interacting protein 1-related protein (Hip1-related). | |||||
|
U2AFM_MOUSE
|
||||||
| NC score | 0.045025 (rank : 93) | θ value | 2.36792 (rank : 83) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 68 | |
| SwissProt Accessions | Q62377 | Gene names | Zrsr2, U2af1-rs2, U2af1l2, U2af1rs2 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 2 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 2) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 2). | |||||
|
U2AFL_HUMAN
|
||||||
| NC score | 0.044681 (rank : 94) | θ value | 3.0926 (rank : 94) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 503 | Shared Neighborhood Hits | 69 | |
| SwissProt Accessions | Q15695, Q13570 | Gene names | ZRSR1, U2AF1-RS1, U2AF1L1, U2AF1RS1, U2AFBPL | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.044633 (rank : 95) | θ value | 5.27518 (rank : 120) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 109 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.044615 (rank : 96) | θ value | 5.27518 (rank : 113) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.044493 (rank : 97) | θ value | 2.36792 (rank : 81) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 111 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
CTGE5_HUMAN
|
||||||
| NC score | 0.044462 (rank : 98) | θ value | 5.27518 (rank : 110) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 800 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | O15320, O00169, Q6P2R8, Q8IX92, Q8IX93 | Gene names | CTAGE5, MEA11, MEA6, MGEA11, MGEA6 | |||
|
Domain Architecture |

|
|||||
| Description | Cutaneous T-cell lymphoma-associated antigen 5 (cTAGE-5 protein) (cTAGE family member 5) (Meningioma-expressed antigen 6/11). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.044079 (rank : 99) | θ value | 4.03905 (rank : 102) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 117 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.043364 (rank : 100) | θ value | 6.88961 (rank : 125) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
TACC3_MOUSE
|
||||||
| NC score | 0.042890 (rank : 101) | θ value | 1.06291 (rank : 53) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 607 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q9JJ11, Q9WVK9 | Gene names | Tacc3, Aint | |||
|
Domain Architecture |

|
|||||
| Description | Transforming acidic coiled-coil-containing protein 3 (ARNT-interacting protein). | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.042807 (rank : 102) | θ value | 0.47712 (rank : 37) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
INCE_MOUSE
|
||||||
| NC score | 0.042322 (rank : 103) | θ value | 8.99809 (rank : 147) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 903 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q9WU62, Q7TN28, Q8BGN4, Q8CGI4 | Gene names | Incenp | |||
|
Domain Architecture |

|
|||||
| Description | Inner centromere protein. | |||||
|
ITSN1_MOUSE
|
||||||
| NC score | 0.041839 (rank : 104) | θ value | 1.81305 (rank : 69) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1669 | Shared Neighborhood Hits | 115 | |
| SwissProt Accessions | Q9Z0R4, Q9R143 | Gene names | Itsn1, Ese1, Itsn | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-1 (EH and SH3 domains protein 1). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.041530 (rank : 105) | θ value | 1.38821 (rank : 54) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
BEGIN_MOUSE
|
||||||
| NC score | 0.041438 (rank : 106) | θ value | 2.36792 (rank : 77) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q68EF6 | Gene names | Begain | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Brain-enriched guanylate kinase-associated protein. | |||||
|
DMD_HUMAN
|
||||||
| NC score | 0.041368 (rank : 107) | θ value | 4.03905 (rank : 98) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | P11532, Q02295, Q14169, Q14170, Q5JYU0, Q7KZ48 | Gene names | DMD | |||
|
Domain Architecture |

|
|||||
| Description | Dystrophin. | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.040464 (rank : 108) | θ value | 1.81305 (rank : 68) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.040175 (rank : 109) | θ value | 2.36792 (rank : 78) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 98 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.039934 (rank : 110) | θ value | 1.38821 (rank : 61) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.039446 (rank : 111) | θ value | 5.27518 (rank : 118) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 110 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
U2AFL_MOUSE
|
||||||
| NC score | 0.039071 (rank : 112) | θ value | 6.88961 (rank : 137) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q64707 | Gene names | Zrsr1, Sp2, Sp2-7, U2af1-rs1, U2af1l1 | |||
|
Domain Architecture |

|
|||||
| Description | U2 small nuclear ribonucleoprotein auxiliary factor 35 kDa subunit- related protein 1 (U2(RNU2) small nuclear RNA auxiliary factor 1-like 1) (CCCH type zinc finger, RNA-binding motif and serine/arginine rich protein 1) (SP2). | |||||
|
CN145_MOUSE
|
||||||
| NC score | 0.038972 (rank : 113) | θ value | 6.88961 (rank : 124) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
CAF1A_MOUSE
|
||||||
| NC score | 0.038781 (rank : 114) | θ value | 1.81305 (rank : 64) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 736 | Shared Neighborhood Hits | 81 | |
| SwissProt Accessions | Q9QWF0 | Gene names | Chaf1a, Caip150 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromatin assembly factor 1 subunit A (CAF-1 subunit A) (Chromatin assembly factor I p150 subunit) (CAF-I 150 kDa subunit) (CAF-Ip150). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.038739 (rank : 115) | θ value | 6.88961 (rank : 132) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 106 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
KMHN1_HUMAN
|
||||||
| NC score | 0.038468 (rank : 116) | θ value | 1.06291 (rank : 52) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 90 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
NUCB2_MOUSE
|
||||||
| NC score | 0.038394 (rank : 117) | θ value | 5.27518 (rank : 119) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
ITSN2_MOUSE
|
||||||
| NC score | 0.038012 (rank : 118) | θ value | 6.88961 (rank : 126) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1371 | Shared Neighborhood Hits | 101 | |
| SwissProt Accessions | Q9Z0R6, Q9Z0R5 | Gene names | Itsn2, Ese2, Sh3d1B | |||
|
Domain Architecture |

|
|||||
| Description | Intersectin-2 (SH3 domain-containing protein 1B) (EH and SH3 domains protein 2) (EH domain and SH3 domain regulator of endocytosis 2). | |||||
|
LIPA3_HUMAN
|
||||||
| NC score | 0.037912 (rank : 119) | θ value | 4.03905 (rank : 99) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.037834 (rank : 120) | θ value | 8.99809 (rank : 142) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 108 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
LIPA3_MOUSE
|
||||||
| NC score | 0.037673 (rank : 121) | θ value | 4.03905 (rank : 100) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 75 | |
| SwissProt Accessions | P60469 | Gene names | Ppfia3 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
GOGA2_MOUSE
|
||||||
| NC score | 0.037642 (rank : 122) | θ value | 8.99809 (rank : 145) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 95 | |
| SwissProt Accessions | Q921M4 | Gene names | Golga2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130). | |||||
|
LMNB1_MOUSE
|
||||||
| NC score | 0.036592 (rank : 123) | θ value | 0.279714 (rank : 31) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 719 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | P14733, Q61791 | Gene names | Lmnb1 | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.036489 (rank : 124) | θ value | 8.99809 (rank : 141) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 107 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
CTRO_MOUSE
|
||||||
| NC score | 0.036302 (rank : 125) | θ value | 0.0736092 (rank : 26) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 2344 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | P49025, O88528, O88937, O88938, Q3UM99, Q8CIJ1 | Gene names | Cit | |||
|
Domain Architecture |

|
|||||
| Description | Citron Rho-interacting kinase (EC 2.7.11.1) (CRIK) (Rho-interacting, serine/threonine-protein kinase 21). | |||||
|
VP13A_HUMAN
|
||||||
| NC score | 0.036158 (rank : 126) | θ value | 4.03905 (rank : 105) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q96RL7, Q5JSX9, Q5JSY0, Q5VYR5, Q702P4, Q709D0, Q86YF8, Q96S61, Q9H995, Q9Y2J1 | Gene names | VPS13A, CHAC, KIAA0986 | |||
|
Domain Architecture |

|
|||||
| Description | Vacuolar protein sorting-associated protein 13A (Chorein) (Chorea- acanthocytosis protein). | |||||
|
PININ_MOUSE
|
||||||
| NC score | 0.035446 (rank : 127) | θ value | 3.0926 (rank : 90) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 777 | Shared Neighborhood Hits | 85 | |
| SwissProt Accessions | O35691, Q8CD89, Q8CGU3 | Gene names | Pnn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pinin. | |||||
|
LMNB1_HUMAN
|
||||||
| NC score | 0.034502 (rank : 128) | θ value | 0.47712 (rank : 36) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 714 | Shared Neighborhood Hits | 83 | |
| SwissProt Accessions | P20700, Q3SYN7, Q96EI6 | Gene names | LMNB1, LMN2, LMNB | |||
|
Domain Architecture |

|
|||||
| Description | Lamin-B1. | |||||
|
LRSM1_HUMAN
|
||||||
| NC score | 0.034410 (rank : 129) | θ value | 0.62314 (rank : 41) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1078 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q6UWE0, Q5VVV0, Q8NB40, Q96GT5, Q96MX5, Q96MZ7 | Gene names | LRSAM1, TAL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin ligase protein LRSAM1 (EC 6.3.2.-) (Leucine-rich repeat and sterile alpha motif-containing protein 1) (Tsg101-associated ligase) (hTAL). | |||||
|
IF2P_HUMAN
|
||||||
| NC score | 0.034369 (rank : 130) | θ value | 3.0926 (rank : 88) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 889 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | O60841, O95805, Q9UF81, Q9UMN7 | Gene names | EIF5B, IF2, KIAA0741 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 5B (eIF-5B) (Translation initiation factor IF-2). | |||||
|
PLEC1_HUMAN
|
||||||
| NC score | 0.033913 (rank : 131) | θ value | 1.38821 (rank : 59) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1326 | Shared Neighborhood Hits | 94 | |
| SwissProt Accessions | Q15149, Q15148, Q16640 | Gene names | PLEC1 | |||
|
Domain Architecture |

|
|||||
| Description | Plectin-1 (PLTN) (PCN) (Hemidesmosomal protein 1) (HD1). | |||||
|
SM1L2_MOUSE
|
||||||
| NC score | 0.033629 (rank : 132) | θ value | 3.0926 (rank : 93) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 93 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
GCP60_MOUSE
|
||||||
| NC score | 0.033164 (rank : 133) | θ value | 8.99809 (rank : 143) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 395 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8BMP6, O35371 | Gene names | Acbd3, Gcp60, Pap7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
KIF5C_MOUSE
|
||||||
| NC score | 0.032838 (rank : 134) | θ value | 8.99809 (rank : 148) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 103 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
BRE1A_MOUSE
|
||||||
| NC score | 0.031878 (rank : 135) | θ value | 5.27518 (rank : 106) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1025 | Shared Neighborhood Hits | 82 | |
| SwissProt Accessions | Q5DTM8, Q3UT10, Q3V350, Q7TT11, Q8BKA8, Q8BKN8, Q8BUF7, Q8BVU4 | Gene names | Rnf20, Bre1a, Kiaa4116 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquitin-protein ligase BRE1A (EC 6.3.2.-) (BRE1-A) (RING finger protein 20). | |||||
|
GCP60_HUMAN
|
||||||
| NC score | 0.031685 (rank : 136) | θ value | 5.27518 (rank : 114) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q9H3P7, Q5VTJ0, Q6P9F1, Q8IZC5, Q8N4D6, Q9H6U3 | Gene names | ACBD3, GCP60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgi resident protein GCP60 (Acyl-CoA-binding domain-containing protein 3) (Golgi phosphoprotein 1) (GOLPH1) (Golgi complex-associated protein 1) (GOCAP1) (PBR- and PKA-associated protein 7) (Peripheral benzodiazepine receptor-associated protein PAP7). | |||||
|
CAPS2_HUMAN
|
||||||
| NC score | 0.029783 (rank : 137) | θ value | 0.279714 (rank : 30) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q86UW7, Q658Q2, Q7Z5T7, Q8IZW9, Q8N7M4, Q9H6P4, Q9HCI1, Q9NWK8 | Gene names | CADPS2, CAPS2, KIAA1591 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 2 (Calcium-dependent activator protein for secretion 2) (CAPS-2). | |||||
|
GBP2_MOUSE
|
||||||
| NC score | 0.029043 (rank : 138) | θ value | 3.0926 (rank : 86) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9Z0E6, Q8CIC6, Q921N2, Q9R1I0 | Gene names | Gbp2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 2 (GTP-binding protein 2) (Guanine nucleotide-binding protein 2) (GBP-2) (mGBP2) (mGBP-2). | |||||
|
CAPS2_MOUSE
|
||||||
| NC score | 0.028922 (rank : 139) | θ value | 0.62314 (rank : 40) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8BYR5, O08903, Q66JM7, Q6PCL7, Q76I88, Q7TMM6, Q80ZV8, Q8BL25, Q8BY04, Q8K3K6 | Gene names | Cadps2, Caps2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium-dependent secretion activator 2 (Calcium-dependent activator protein for secretion 2) (CAPS-2). | |||||
|
PKHG1_HUMAN
|
||||||
| NC score | 0.028218 (rank : 140) | θ value | 0.47712 (rank : 38) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9ULL1, Q5T1F2 | Gene names | PLEKHG1, KIAA1209 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Pleckstrin homology domain-containing family G member 1. | |||||
|
MYO5A_MOUSE
|
||||||
| NC score | 0.027838 (rank : 141) | θ value | 1.81305 (rank : 70) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 104 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
TF2H1_MOUSE
|
||||||
| NC score | 0.025839 (rank : 142) | θ value | 1.81305 (rank : 76) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBA9, O35637 | Gene names | Gtf2h1 | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex p62 subunit (Basic transcription factor 62 kDa subunit) (BTF2-p62) (General transcription factor IIH polypeptide 1). | |||||
|
TF2H1_HUMAN
|
||||||
| NC score | 0.025835 (rank : 143) | θ value | 1.81305 (rank : 75) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P32780, Q9H5K5, Q9NQD9 | Gene names | GTF2H1, BTF2 | |||
|
Domain Architecture |

|
|||||
| Description | TFIIH basal transcription factor complex p62 subunit (Basic transcription factor 62 kDa subunit) (BTF2-p62) (General transcription factor IIH polypeptide 1). | |||||
|
GBP1_HUMAN
|
||||||
| NC score | 0.024880 (rank : 144) | θ value | 1.81305 (rank : 67) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 339 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P32455 | Gene names | GBP1 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 1 (GTP-binding protein 1) (Guanine nucleotide-binding protein 1) (GBP-1) (HuGBP-1). | |||||
|
REN3A_HUMAN
|
||||||
| NC score | 0.024840 (rank : 145) | θ value | 8.99809 (rank : 158) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 248 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | Q9H1J1, Q5T8C3, Q5T8C9, Q7Z6N3, Q86YK1, Q9BZI8 | Gene names | UPF3A, RENT3A, UPF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Regulator of nonsense transcripts 3A (Nonsense mRNA reducing factor 3A) (Up-frameshift suppressor 3 homolog A) (hUpf3). | |||||
|
RNF8_MOUSE
|
||||||
| NC score | 0.024161 (rank : 146) | θ value | 3.0926 (rank : 92) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 618 | Shared Neighborhood Hits | 71 | |
| SwissProt Accessions | Q8VC56, Q9JK13 | Gene names | Rnf8 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin ligase protein RNF8 (EC 6.3.2.-) (RING finger protein 8) (AIP37) (ActA-interacting protein 37) (LaXp180). | |||||
|
RPGR1_MOUSE
|
||||||
| NC score | 0.023536 (rank : 147) | θ value | 6.88961 (rank : 134) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 713 | Shared Neighborhood Hits | 67 | |
| SwissProt Accessions | Q9EPQ2, Q8CAC2, Q8CDJ9, Q91WE0, Q9CUK6, Q9D5Q1 | Gene names | Rpgrip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | X-linked retinitis pigmentosa GTPase regulator-interacting protein 1 (RPGR-interacting protein 1). | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.023399 (rank : 148) | θ value | 2.36792 (rank : 79) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 114 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
MYO5B_HUMAN
|
||||||
| NC score | 0.022677 (rank : 149) | θ value | 5.27518 (rank : 117) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 911 | Shared Neighborhood Hits | 97 | |
| SwissProt Accessions | Q9ULV0 | Gene names | MYO5B, KIAA1119 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5B (Myosin Vb). | |||||
|
MAT1_MOUSE
|
||||||
| NC score | 0.021775 (rank : 150) | θ value | 4.03905 (rank : 101) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P51949, Q3TZP0, Q9D8A0, Q9D8D2 | Gene names | Mnat1, Mat1 | |||
|
Domain Architecture |

|
|||||
| Description | CDK-activating kinase assembly factor MAT1 (RING finger protein MAT1) (Menage a trois) (CDK7/cyclin H assembly factor) (p36) (p35). | |||||
|
MICA1_MOUSE
|
||||||
| NC score | 0.021759 (rank : 151) | θ value | 3.0926 (rank : 89) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8VDP3 | Gene names | Mical1, Mical, Nical | |||
|
Domain Architecture |

|
|||||
| Description | NEDD9-interacting protein with calponin homology and LIM domains (Molecule interacting with CasL protein 1). | |||||
|
MAT1_HUMAN
|
||||||
| NC score | 0.020631 (rank : 152) | θ value | 5.27518 (rank : 116) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P51948, Q15817 | Gene names | MNAT1, CAP35, MAT1, RNF66 | |||
|
Domain Architecture |

|
|||||
| Description | CDK-activating kinase assembly factor MAT1 (RING finger protein MAT1) (Menage a trois) (CDK7/cyclin H assembly factor) (p36) (p35) (Cyclin G1-interacting protein) (RING finger protein 66). | |||||
|
MOES_HUMAN
|
||||||
| NC score | 0.020462 (rank : 153) | θ value | 8.99809 (rank : 149) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 892 | Shared Neighborhood Hits | 91 | |
| SwissProt Accessions | P26038 | Gene names | MSN | |||
|
Domain Architecture |

|
|||||
| Description | Moesin (Membrane-organizing extension spike protein). | |||||
|
RN168_HUMAN
|
||||||
| NC score | 0.019897 (rank : 154) | θ value | 1.38821 (rank : 60) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q8IYW5, Q8NA67, Q96NS4 | Gene names | RNF168 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RING finger protein 168. | |||||
|
MYO6_HUMAN
|
||||||
| NC score | 0.018593 (rank : 155) | θ value | 8.99809 (rank : 150) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 84 | |
| SwissProt Accessions | Q9UM54, Q9BZZ7, Q9UEG2 | Gene names | MYO6, KIAA0389 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-6 (Myosin VI). | |||||
|
FYB_MOUSE
|
||||||
| NC score | 0.018106 (rank : 156) | θ value | 1.81305 (rank : 66) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | O35601, Q9Z2H3 | Gene names | Fyb | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
OXR1_MOUSE
|
||||||
| NC score | 0.017850 (rank : 157) | θ value | 1.38821 (rank : 57) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 516 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q4KMM3, Q5FWW1, Q99L06, Q99MK1, Q99MP4 | Gene names | Oxr1, C7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oxidation resistance protein 1 (Protein C7). | |||||
|
UBXD8_HUMAN
|
||||||
| NC score | 0.016622 (rank : 158) | θ value | 5.27518 (rank : 121) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 131 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96CS3, O94963, Q8IUF2, Q9BRP2, Q9BVM7 | Gene names | UBXD8, ETEA, KIAA0887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 8 (Protein ETEA). | |||||
|
UBXD8_MOUSE
|
||||||
| NC score | 0.016076 (rank : 159) | θ value | 6.88961 (rank : 138) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 133 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q3TDN2, Q3U9W2, Q80TP7, Q80W42 | Gene names | Ubxd8, Kiaa0887 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | UBX domain-containing protein 8. | |||||
|
TAF3_MOUSE
|
||||||
| NC score | 0.015121 (rank : 160) | θ value | 8.99809 (rank : 160) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 463 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q5HZG4, Q3U490, Q3UWX2, Q8BIU8, Q99JH4 | Gene names | Taf3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
RAB1B_MOUSE
|
||||||
| NC score | 0.014547 (rank : 161) | θ value | 8.99809 (rank : 156) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9D1G1, Q3U0N1 | Gene names | Rab1b | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-1B. | |||||
|
RAB1B_HUMAN
|
||||||
| NC score | 0.014530 (rank : 162) | θ value | 8.99809 (rank : 155) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 295 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9H0U4 | Gene names | RAB1B | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-1B. | |||||
|
RAB15_HUMAN
|
||||||
| NC score | 0.013916 (rank : 163) | θ value | 3.0926 (rank : 91) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 283 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P59190, Q86TX7, Q8IW89 | Gene names | RAB15 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-15. | |||||
|
CECR2_HUMAN
|
||||||
| NC score | 0.013461 (rank : 164) | θ value | 1.81305 (rank : 65) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q9BXF3, Q658Z4, Q96P58, Q9C0C3 | Gene names | CECR2, KIAA1740 | |||
|
Domain Architecture |

|
|||||
| Description | Cat eye syndrome critical region protein 2. | |||||
|
FYB_HUMAN
|
||||||
| NC score | 0.013372 (rank : 165) | θ value | 5.27518 (rank : 112) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15117, O00359 | Gene names | FYB, SLAP130 | |||
|
Domain Architecture |

|
|||||
| Description | FYN-binding protein (FYN-T-binding protein) (FYB-120/130) (p120/p130) (SLP-76-associated phosphoprotein) (SLAP-130). | |||||
|
RAB1A_HUMAN
|
||||||
| NC score | 0.012891 (rank : 166) | θ value | 8.99809 (rank : 153) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P62820, P11476, Q96N61, Q9Y3T2 | Gene names | RAB1A, RAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-related protein Rab-1A (YPT1-related protein). | |||||
|
RAB1A_MOUSE
|
||||||
| NC score | 0.012891 (rank : 167) | θ value | 8.99809 (rank : 154) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 292 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P62821, P11476, Q3TX44, Q811M4, Q96N61, Q9Y3T2 | Gene names | Rab1A, Rab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ras-related protein Rab-1A (YPT1-related protein). | |||||
|
DBP_MOUSE
|
||||||
| NC score | 0.011745 (rank : 168) | θ value | 4.03905 (rank : 96) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 137 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q60925, Q8VCX3 | Gene names | Dbp | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein). | |||||
|
MEFV_HUMAN
|
||||||
| NC score | 0.011122 (rank : 169) | θ value | 1.38821 (rank : 56) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | O15553, Q96PN4, Q96PN5 | Gene names | MEFV, MEF | |||
|
Domain Architecture |

|
|||||
| Description | Pyrin (Marenostrin). | |||||
|
DBP_HUMAN
|
||||||
| NC score | 0.010936 (rank : 170) | θ value | 5.27518 (rank : 111) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q10586 | Gene names | DBP | |||
|
Domain Architecture |

|
|||||
| Description | D site-binding protein (Albumin D box-binding protein) (Albumin D- element-binding protein) (TAXREB302). | |||||
|
RAB10_HUMAN
|
||||||
| NC score | 0.010765 (rank : 171) | θ value | 4.03905 (rank : 103) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P61026, O88386, Q6IA52, Q9D7X6, Q9H0T3 | Gene names | RAB10 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-10. | |||||
|
RAB10_MOUSE
|
||||||
| NC score | 0.010765 (rank : 172) | θ value | 4.03905 (rank : 104) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 278 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P61027, O88386, Q9D7X6 | Gene names | Rab10 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-10. | |||||
|
RAB15_MOUSE
|
||||||
| NC score | 0.010680 (rank : 173) | θ value | 6.88961 (rank : 131) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 284 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q8K386 | Gene names | Rab15 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-15. | |||||
|
TRI32_MOUSE
|
||||||
| NC score | 0.010653 (rank : 174) | θ value | 2.36792 (rank : 82) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8CH72, Q8K055 | Gene names | Trim32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-). | |||||
|
BRD3_MOUSE
|
||||||
| NC score | 0.010264 (rank : 175) | θ value | 6.88961 (rank : 123) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 402 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q8K2F0, Q8C665, Q8CAX7, Q9JI25 | Gene names | Brd3, Fsrg2 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 3 (Bromodomain-containing FSH-like protein FSRG2). | |||||
|
RAB13_HUMAN
|
||||||
| NC score | 0.008146 (rank : 176) | θ value | 8.99809 (rank : 152) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 286 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P51153, Q96GU4 | Gene names | RAB13 | |||
|
Domain Architecture |

|
|||||
| Description | Ras-related protein Rab-13. | |||||
|
MINK1_MOUSE
|
||||||
| NC score | 0.007681 (rank : 177) | θ value | 0.62314 (rank : 43) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1451 | Shared Neighborhood Hits | 73 | |
| SwissProt Accessions | Q9JM52, Q921M6, Q9JM92 | Gene names | Mink1, Map4k6, Mink | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
MINK1_HUMAN
|
||||||
| NC score | 0.007662 (rank : 178) | θ value | 0.62314 (rank : 42) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 74 | |
| SwissProt Accessions | Q8N4C8, Q9P1X1, Q9P2R8 | Gene names | MINK1, MAP4K6, MINK | |||
|
Domain Architecture |

|
|||||
| Description | Misshapen-like kinase 1 (EC 2.7.11.1) (Mitogen-activated protein kinase kinase kinase kinase 6) (MAPK/ERK kinase kinase kinase 6) (MEK kinase kinase 6) (MEKKK 6) (Misshapen/NIK-related kinase) (GCK family kinase MiNK). | |||||
|
TWST2_MOUSE
|
||||||
| NC score | 0.007296 (rank : 179) | θ value | 6.88961 (rank : 136) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9D030 | Gene names | Twist2, Dermo1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
TWST2_HUMAN
|
||||||
| NC score | 0.007281 (rank : 180) | θ value | 6.88961 (rank : 135) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8WVJ9 | Gene names | TWIST2, DERMO1 | |||
|
Domain Architecture |

|
|||||
| Description | Twist-related protein 2 (Dermis expressed protein 1) (Dermo-1). | |||||
|
PTN2_MOUSE
|
||||||
| NC score | 0.006322 (rank : 181) | θ value | 8.99809 (rank : 151) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q06180, Q3V259, Q922E7 | Gene names | Ptpn2, Ptpt | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type 2 (EC 3.1.3.48) (Protein-tyrosine phosphatase PTP-2) (MPTP). | |||||
|
GDIT_HUMAN
|
||||||
| NC score | 0.004579 (rank : 182) | θ value | 8.99809 (rank : 144) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99819, Q4TT69, Q96S29 | Gene names | ARHGDIG | |||
|
Domain Architecture |

|
|||||
| Description | Rho GDP-dissociation inhibitor 3 (Rho GDI 3) (Rho-GDI gamma). | |||||
|
SUV91_MOUSE
|
||||||
| NC score | 0.004491 (rank : 183) | θ value | 8.99809 (rank : 159) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 69 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O54864, Q9JLC7, Q9JLP8 | Gene names | Suv39h1, Suv39h | |||
|
Domain Architecture |

|
|||||
| Description | Histone-lysine N-methyltransferase, H3 lysine-9 specific 1 (EC 2.1.1.43) (Histone H3-K9 methyltransferase 1) (H3-K9-HMTase 1) (Suppressor of variegation 3-9 homolog 1) (Su(var)3-9 homolog 1) (Position-effect variegation 3-9 homolog). | |||||
|
UBP37_HUMAN
|
||||||
| NC score | 0.003453 (rank : 184) | θ value | 8.99809 (rank : 161) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q86T82, Q9HCH8 | Gene names | USP37, KIAA1594 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 37 (EC 3.1.2.15) (Ubiquitin thioesterase 37) (Ubiquitin-specific-processing protease 37) (Deubiquitinating enzyme 37). | |||||
|
CD2L1_MOUSE
|
||||||
| NC score | 0.002846 (rank : 185) | θ value | 5.27518 (rank : 108) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1463 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P24788, Q3UI03, Q61399, Q7TST4, Q8BP53 | Gene names | Cdc2l1 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1). | |||||
|
NAL13_HUMAN
|
||||||
| NC score | 0.002554 (rank : 186) | θ value | 6.88961 (rank : 128) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q86W25, Q7RTR5 | Gene names | NALP13, NOD14 | |||
|
Domain Architecture |

|
|||||
| Description | NACHT, LRR and PYD-containing protein 13 (Nucleotide-binding oligomerization domain protein 14). | |||||
|
CD2L1_HUMAN
|
||||||
| NC score | 0.002507 (rank : 187) | θ value | 8.99809 (rank : 140) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 1467 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P21127, O95265, Q12817, Q12818, Q12819, Q12820, Q12822, Q8N530, Q9NZS5, Q9UBJ0, Q9UBQ1, Q9UBR0, Q9UNY2, Q9UP57, Q9UP58, Q9UP59 | Gene names | CDC2L1, PITSLREA, PK58 | |||
|
Domain Architecture |

|
|||||
| Description | PITSLRE serine/threonine-protein kinase CDC2L1 (EC 2.7.11.22) (Galactosyltransferase-associated protein kinase p58/GTA) (Cell division cycle 2-like protein kinase 1) (CLK-1) (CDK11) (p58 CLK-1). | |||||
|
PP16A_MOUSE
|
||||||
| NC score | -0.000514 (rank : 188) | θ value | 6.88961 (rank : 130) | |||
| Query Neighborhood Hits | 161 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q923M0, Q4V9Z1 | Gene names | Ppp1r16a, Mypt3 | |||
|
Domain Architecture |

|
|||||
| Description | Protein phosphatase 1 regulatory subunit 16A (Myosin phosphatase targeting subunit 3). | |||||