Please be patient as the page loads
|
SC15B_HUMAN
|
||||||
| SwissProt Accessions | Q9Y2D4 | Gene names | SEC15L2, KIAA0919, SEC15B | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component Sec15B. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
EXOC6_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.966476 (rank : 3) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TAG9, Q9NZ24 | Gene names | EXOC6, SEC15A, SEC15L, SEC15L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 6 (Exocyst complex component Sec15A) (Sec15- like 1). | |||||
|
EXOC6_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.972728 (rank : 2) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R313 | Gene names | Exoc6, Sec15a, Sec15l1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 6 (Exocyst complex component Sec15A) (Sec15- like 1). | |||||
|
SC15B_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y2D4 | Gene names | SEC15L2, KIAA0919, SEC15B | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component Sec15B. | |||||
|
CE290_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 4) | NC score | 0.079078 (rank : 7) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 5) | NC score | 0.066271 (rank : 18) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 6) | NC score | 0.076575 (rank : 8) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 0.0563607 (rank : 7) | NC score | 0.066589 (rank : 16) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 0.163984 (rank : 8) | NC score | 0.054851 (rank : 84) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
ANGL3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 9) | NC score | 0.039471 (rank : 179) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 32 | |
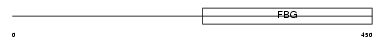
| SwissProt Accessions | Q9Y5C1 | Gene names | ANGPTL3, ANGPT5 | |||
|
Domain Architecture |
|
|||||
| Description | Angiopoietin-related protein 3 precursor (Angiopoietin-like 3) (Angiopoietin-5) (ANG-5). | |||||
|
KTN1_HUMAN
|
||||||
| θ value | 0.279714 (rank : 10) | NC score | 0.096861 (rank : 5) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
LIPA2_MOUSE
|
||||||
| θ value | 0.279714 (rank : 11) | NC score | 0.068397 (rank : 11) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
PEPL_MOUSE
|
||||||
| θ value | 0.279714 (rank : 12) | NC score | 0.065132 (rank : 21) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
SPC25_HUMAN
|
||||||
| θ value | 0.279714 (rank : 13) | NC score | 0.102527 (rank : 4) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HBM1 | Gene names | SPBC25, SPC25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc25 (hSpc25). | |||||
|
YO11_MOUSE
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.083329 (rank : 6) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P11260 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Hypothetical protein ORF-1137. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.62314 (rank : 15) | NC score | 0.063224 (rank : 26) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
KIF5C_HUMAN
|
||||||
| θ value | 0.62314 (rank : 16) | NC score | 0.054960 (rank : 80) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
MYH3_HUMAN
|
||||||
| θ value | 0.62314 (rank : 17) | NC score | 0.053764 (rank : 109) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
ROCK1_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.031379 (rank : 182) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
SMC1A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.064588 (rank : 23) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
KINH_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.057308 (rank : 56) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
RAD50_HUMAN
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.067938 (rank : 13) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
LIPA2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 22) | NC score | 0.067584 (rank : 14) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
CING_HUMAN
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.062249 (rank : 29) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
NEBL_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.048217 (rank : 177) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O76041, Q9UIC4 | Gene names | NEBL | |||
|
Domain Architecture |

|
|||||
| Description | Nebulette (Actin-binding Z-disk protein). | |||||
|
TPR_HUMAN
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.062091 (rank : 30) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
ANGP2_HUMAN
|
||||||
| θ value | 1.81305 (rank : 26) | NC score | 0.033906 (rank : 180) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 42 | |
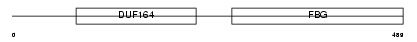
| SwissProt Accessions | O15123, Q9NRR7, Q9P2Y7 | Gene names | ANGPT2 | |||
|
Domain Architecture |
|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
IFT81_HUMAN
|
||||||
| θ value | 1.81305 (rank : 27) | NC score | 0.068169 (rank : 12) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
KINH_MOUSE
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.056204 (rank : 70) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
LAMB1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.032522 (rank : 181) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
MRCKB_HUMAN
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.028941 (rank : 184) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
NASP_HUMAN
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.057911 (rank : 49) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
IF3A_HUMAN
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.064315 (rank : 24) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
IFT81_MOUSE
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.066331 (rank : 17) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
SMC1A_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.062906 (rank : 28) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
B4GT6_MOUSE
|
||||||
| θ value | 3.0926 (rank : 35) | NC score | 0.013219 (rank : 190) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVK5 | Gene names | B4galt6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
CA065_HUMAN
|
||||||
| θ value | 3.0926 (rank : 36) | NC score | 0.054598 (rank : 91) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N715, Q8N746, Q8NA93 | Gene names | C1orf65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf65. | |||||
|
CR034_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.071233 (rank : 9) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5BJE1, Q6ZP67, Q6ZU20 | Gene names | C18orf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34. | |||||
|
CYTSA_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.058157 (rank : 46) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
IF3A_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.063137 (rank : 27) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
BPAEA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.056049 (rank : 73) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CYTSA_MOUSE
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.058899 (rank : 44) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
EEA1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.059720 (rank : 41) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
GRAP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.055773 (rank : 77) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
HAT1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 44) | NC score | 0.050852 (rank : 166) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14929, Q49A44, Q53QF0, Q53SU4 | Gene names | HAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase type B catalytic subunit (EC 2.3.1.48). | |||||
|
LIPA3_HUMAN
|
||||||
| θ value | 4.03905 (rank : 45) | NC score | 0.065275 (rank : 19) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
LIPA3_MOUSE
|
||||||
| θ value | 4.03905 (rank : 46) | NC score | 0.061861 (rank : 32) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P60469 | Gene names | Ppfia3 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.052304 (rank : 141) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.050650 (rank : 169) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.052217 (rank : 144) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
TPM4_MOUSE
|
||||||
| θ value | 4.03905 (rank : 50) | NC score | 0.052735 (rank : 130) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
FLII_HUMAN
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.004918 (rank : 194) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13045 | Gene names | FLII, FLIL | |||
|
Domain Architecture |

|
|||||
| Description | Protein flightless-1 homolog. | |||||
|
KIF5A_MOUSE
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.051667 (rank : 154) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
TAOK3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.018234 (rank : 186) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1494 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H2K8, Q658N1, Q8IUM4, Q9HC79, Q9NZM9, Q9UHG7 | Gene names | TAOK3, DPK, JIK, KDS, MAP3K18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3) (Jun kinase-inhibitory kinase) (JNK/SAPK- inhibitory kinase) (Dendritic cell-derived protein kinase) (Cutaneous T-cell lymphoma tumor antigen HD-CL-09) (CTCL tumor antigen HD-CL-09) (Kinase from chicken homolog A) (hKFC-A). | |||||
|
UACA_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.050656 (rank : 168) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
CETP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.017833 (rank : 187) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
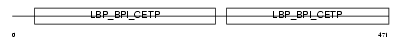
| SwissProt Accessions | P11597, Q13987, Q13988 | Gene names | CETP | |||
|
Domain Architecture |
|
|||||
| Description | Cholesteryl ester transfer protein precursor (Lipid transfer protein I). | |||||
|
DSPP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.009768 (rank : 191) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
ERC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.056400 (rank : 68) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
HOOK1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.053288 (rank : 117) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
MYH8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.052182 (rank : 146) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
SYCP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.067254 (rank : 15) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TAOK3_MOUSE
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.016516 (rank : 188) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1458 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BYC6, Q3V3B3 | Gene names | Taok3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3). | |||||
|
CHD9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 62) | NC score | 0.004524 (rank : 195) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||
|
CN108_MOUSE
|
||||||
| θ value | 8.99809 (rank : 63) | NC score | 0.013228 (rank : 189) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
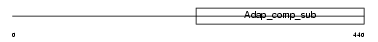
| SwissProt Accessions | Q8BJ63, Q3TPT5, Q8BII2, Q8BJ31, Q8BJ58, Q8BY11, Q8R5B1 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Protein C14orf108 homolog. | |||||
|
DYH5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 64) | NC score | 0.019033 (rank : 185) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
GOGA5_HUMAN
|
||||||
| θ value | 8.99809 (rank : 65) | NC score | 0.054475 (rank : 95) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
LAMA4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 66) | NC score | 0.029619 (rank : 183) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P97927, O88785, P70409 | Gene names | Lama4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
LRRF2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 67) | NC score | 0.059732 (rank : 40) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
MY18B_HUMAN
|
||||||
| θ value | 8.99809 (rank : 68) | NC score | 0.047635 (rank : 178) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
PARD3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 69) | NC score | 0.007799 (rank : 193) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
REST_MOUSE
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.057389 (rank : 55) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
TRIM6_MOUSE
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.008017 (rank : 192) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGE7, Q99PQ6 | Gene names | Trim6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 6. | |||||
|
ZN365_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.055695 (rank : 78) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BG89, Q80TQ4, Q8BK39, Q8BXT2 | Gene names | Znf365, Kiaa0844, Zfp365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365 (Su48). | |||||
|
3BP5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.050462 (rank : 172) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Z131, Q8C903, Q8VCZ0 | Gene names | Sh3bp5, Sab | |||
|
Domain Architecture |
|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
AKAP9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.055132 (rank : 79) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
APOA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.057017 (rank : 60) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P06728 | Gene names | Apoa4 | |||
|
Domain Architecture |
|
|||||
| Description | Apolipoprotein A-IV precursor (Apo-AIV) (ApoA-IV). | |||||
|
BICD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.053005 (rank : 123) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
BICD1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.053824 (rank : 106) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
BICD2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.051489 (rank : 157) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
C102B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.054510 (rank : 94) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q68D86, Q7Z467, Q8NDK7, Q9H5C1 | Gene names | CCDC102B, C18orf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 102B. | |||||
|
CALD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.052165 (rank : 147) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CCD11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.050737 (rank : 167) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D439 | Gene names | Ccdc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
CCD13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.052466 (rank : 137) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
CCD19_HUMAN
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.051268 (rank : 159) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
CCD27_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.051535 (rank : 156) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
CCD39_HUMAN
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.060196 (rank : 36) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD39_MOUSE
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.057929 (rank : 48) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CCD41_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.052165 (rank : 148) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
CCD41_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.053430 (rank : 114) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
CCD46_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.054861 (rank : 82) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CCD46_MOUSE
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.054140 (rank : 102) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
CCD62_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.065137 (rank : 20) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6P9F0, Q6ZVF2, Q86VJ0, Q9BYZ5 | Gene names | CCDC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60 (Aaa-protein) (Protein TSP- NY). | |||||
|
CCHCR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.050231 (rank : 174) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
CE152_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.059279 (rank : 43) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CENPF_HUMAN
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.057758 (rank : 51) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CEP55_MOUSE
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.050972 (rank : 163) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
CEP63_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.054841 (rank : 85) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
CEP70_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.064250 (rank : 25) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NHQ1, Q96B31, Q9H2C3, Q9H2Z1, Q9H940 | Gene names | CEP70, BITE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 70 kDa (Cep70 protein) (p10-binding protein). | |||||
|
CEP70_MOUSE
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.052244 (rank : 142) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6IQY5, Q9CRL9, Q9CTS4, Q9JIC1 | Gene names | Cep70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 70 kDa (Cep70 protein). | |||||
|
CF060_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.054857 (rank : 83) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
CI039_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.060619 (rank : 34) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
CI093_HUMAN
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.056006 (rank : 74) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
CING_MOUSE
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.056472 (rank : 66) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
CJ078_MOUSE
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.064756 (rank : 22) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
CJ118_HUMAN
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.052694 (rank : 132) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
CJ118_MOUSE
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.053822 (rank : 108) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
CK5P2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.050944 (rank : 164) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
CK5P2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.052969 (rank : 124) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
CN145_HUMAN
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.053120 (rank : 119) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
CN145_MOUSE
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.053824 (rank : 107) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
CP135_HUMAN
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.056665 (rank : 63) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CP135_MOUSE
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.058089 (rank : 47) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CP250_HUMAN
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.053384 (rank : 115) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
CP250_MOUSE
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.053599 (rank : 113) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CR034_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.061820 (rank : 33) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
CROCC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.053049 (rank : 122) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
CYLN2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.051115 (rank : 161) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
CYLN2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.051010 (rank : 162) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
DESP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.054395 (rank : 96) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
EEA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.057041 (rank : 59) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
ERC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.056477 (rank : 65) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
EVPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.052746 (rank : 129) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
EVPL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.052691 (rank : 133) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.052212 (rank : 145) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.052796 (rank : 127) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
GAS8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.056272 (rank : 69) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
GAS8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.054163 (rank : 101) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q60779, Q99L71 | Gene names | Gas8, Gas11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.059814 (rank : 39) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
GOGA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.052372 (rank : 139) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.054529 (rank : 93) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
GOGA3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.054573 (rank : 92) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
GOGA3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.054723 (rank : 87) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
GOGA4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.056414 (rank : 67) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
GOGA5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.052239 (rank : 143) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
GOGA6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.051279 (rank : 158) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.056966 (rank : 61) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
GRAP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.052954 (rank : 125) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
HMMR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.054239 (rank : 99) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
HMMR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.055882 (rank : 75) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
IFT74_HUMAN
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.056138 (rank : 72) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
IFT74_MOUSE
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.054789 (rank : 86) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
INVO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.051124 (rank : 160) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
K0555_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.054250 (rank : 98) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
KIF5C_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.051830 (rank : 151) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
KMHN1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.054206 (rank : 100) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
KMHN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.054008 (rank : 103) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
KNTC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.054651 (rank : 89) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
KNTC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.054270 (rank : 97) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
KTN1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.061993 (rank : 31) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
LIPA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.060020 (rank : 37) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
LIPB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.050223 (rank : 175) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8C8U0, Q69ZN5, Q6GQV3, Q80VB4, Q9CUT7 | Gene names | Ppfibp1, Kiaa1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1). | |||||
|
LRRF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.051822 (rank : 152) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q91WK0, Q8BVD1, Q9CU89 | Gene names | Lrrfip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
MD1L1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.050520 (rank : 171) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
MD1L1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.053861 (rank : 105) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
MYH1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.052402 (rank : 138) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
MYH6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.052678 (rank : 134) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
MYH7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.050600 (rank : 170) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.050297 (rank : 173) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
NIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.055828 (rank : 76) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
NIN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.053615 (rank : 112) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
NUF2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.054608 (rank : 90) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
NUF2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.050920 (rank : 165) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99P69, Q8BTK7, Q8VE05, Q9CST5 | Gene names | Cdca1, Nuf2r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (Cell division cycle-associated protein 1). | |||||
|
NUMA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.059374 (rank : 42) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
OFD1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.051643 (rank : 155) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
OPTN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.052899 (rank : 126) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
OPTN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.051891 (rank : 150) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
PEPL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.057083 (rank : 58) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
PIBF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.056547 (rank : 64) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
RABE1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.052371 (rank : 140) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
RABE1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.053862 (rank : 104) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
RAD50_MOUSE
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.060611 (rank : 35) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.054904 (rank : 81) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
RB6I2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.057802 (rank : 50) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
RBCC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.057126 (rank : 57) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.053616 (rank : 111) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.059898 (rank : 38) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
REST_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.057557 (rank : 53) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
SAS6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.052776 (rank : 128) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6UVJ0, Q8N3K0 | Gene names | SASS6, SAS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog (HsSAS-6). | |||||
|
SAS6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.053096 (rank : 120) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
SDCG8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.052500 (rank : 136) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
SM1L2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.058847 (rank : 45) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
SM1L2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 181) | NC score | 0.056917 (rank : 62) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
SMC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 182) | NC score | 0.052523 (rank : 135) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
SMC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 183) | NC score | 0.053230 (rank : 118) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
SMC3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 184) | NC score | 0.051722 (rank : 153) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
SMC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 185) | NC score | 0.052162 (rank : 149) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
SMC4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 186) | NC score | 0.053314 (rank : 116) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
SMC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 187) | NC score | 0.052733 (rank : 131) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
SPC25_MOUSE
|
||||||
| θ value | θ > 10 (rank : 188) | NC score | 0.053694 (rank : 110) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3UA16, Q9D021, Q9D1K6 | Gene names | Spbc25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc25. | |||||
|
TAXB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 189) | NC score | 0.053088 (rank : 121) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
TAXB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 190) | NC score | 0.054707 (rank : 88) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
TEX9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 191) | NC score | 0.050064 (rank : 176) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8N6V9 | Gene names | TEX9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein. | |||||
|
TMF1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 192) | NC score | 0.056165 (rank : 71) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
TRAIP_HUMAN
|
||||||
| θ value | θ > 10 (rank : 193) | NC score | 0.068871 (rank : 10) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
TRHY_HUMAN
|
||||||
| θ value | θ > 10 (rank : 194) | NC score | 0.057584 (rank : 52) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
TRIPB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 195) | NC score | 0.057512 (rank : 54) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
SC15B_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 72 | Shared Neighborhood Hits | 72 | |
| SwissProt Accessions | Q9Y2D4 | Gene names | SEC15L2, KIAA0919, SEC15B | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component Sec15B. | |||||
|
EXOC6_MOUSE
|
||||||
| NC score | 0.972728 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 26 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8R313 | Gene names | Exoc6, Sec15a, Sec15l1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 6 (Exocyst complex component Sec15A) (Sec15- like 1). | |||||
|
EXOC6_HUMAN
|
||||||
| NC score | 0.966476 (rank : 3) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8TAG9, Q9NZ24 | Gene names | EXOC6, SEC15A, SEC15L, SEC15L1 | |||
|
Domain Architecture |

|
|||||
| Description | Exocyst complex component 6 (Exocyst complex component Sec15A) (Sec15- like 1). | |||||
|
SPC25_HUMAN
|
||||||
| NC score | 0.102527 (rank : 4) | θ value | 0.279714 (rank : 13) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | Q9HBM1 | Gene names | SPBC25, SPC25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc25 (hSpc25). | |||||
|
KTN1_HUMAN
|
||||||
| NC score | 0.096861 (rank : 5) | θ value | 0.279714 (rank : 10) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1197 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q86UP2, Q13999, Q14707, Q15387, Q86W57 | Gene names | KTN1, CG1, KIAA0004 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin (Kinesin receptor) (CG-1 antigen). | |||||
|
YO11_MOUSE
|
||||||
| NC score | 0.083329 (rank : 6) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 38 | |
| SwissProt Accessions | P11260 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Hypothetical protein ORF-1137. | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.079078 (rank : 7) | θ value | 0.0252991 (rank : 4) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.076575 (rank : 8) | θ value | 0.0252991 (rank : 6) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
CR034_HUMAN
|
||||||
| NC score | 0.071233 (rank : 9) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 538 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q5BJE1, Q6ZP67, Q6ZU20 | Gene names | C18orf34 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34. | |||||
|
TRAIP_HUMAN
|
||||||
| NC score | 0.068871 (rank : 10) | θ value | θ > 10 (rank : 193) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 780 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9BWF2, O00467 | Gene names | TRAIP, TRIP | |||
|
Domain Architecture |

|
|||||
| Description | TRAF-interacting protein. | |||||
|
LIPA2_MOUSE
|
||||||
| NC score | 0.068397 (rank : 11) | θ value | 0.279714 (rank : 11) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1282 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8BSS9, Q8BN73 | Gene names | Ppfia2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
IFT81_HUMAN
|
||||||
| NC score | 0.068169 (rank : 12) | θ value | 1.81305 (rank : 27) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8WYA0, Q8NB51, Q9BSV2, Q9UNY8 | Gene names | IFT81, CDV1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
RAD50_HUMAN
|
||||||
| NC score | 0.067938 (rank : 13) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q92878, O43254, Q6GMT7, Q6P5X3, Q9UP86 | Gene names | RAD50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (hRAD50). | |||||
|
LIPA2_HUMAN
|
||||||
| NC score | 0.067584 (rank : 14) | θ value | 1.06291 (rank : 22) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1044 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O75334 | Gene names | PPFIA2 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-2 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-2) (PTPRF-interacting protein alpha-2). | |||||
|
SYCP1_HUMAN
|
||||||
| NC score | 0.067254 (rank : 15) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q15431, O14963 | Gene names | SYCP1, SCP1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.066589 (rank : 16) | θ value | 0.0563607 (rank : 7) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.066331 (rank : 17) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.066271 (rank : 18) | θ value | 0.0252991 (rank : 5) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
LIPA3_HUMAN
|
||||||
| NC score | 0.065275 (rank : 19) | θ value | 4.03905 (rank : 45) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 828 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O75145, Q9H8B5, Q9UEW4 | Gene names | PPFIA3, KIAA0654 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
CCD62_HUMAN
|
||||||
| NC score | 0.065137 (rank : 20) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6P9F0, Q6ZVF2, Q86VJ0, Q9BYZ5 | Gene names | CCDC62 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 60 (Aaa-protein) (Protein TSP- NY). | |||||
|
PEPL_MOUSE
|
||||||
| NC score | 0.065132 (rank : 21) | θ value | 0.279714 (rank : 12) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q9R269, O70231, Q9CUT1, Q9JLZ7 | Gene names | Ppl | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin. | |||||
|
CJ078_MOUSE
|
||||||
| NC score | 0.064756 (rank : 22) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 559 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BP27, Q3UIQ6, Q8R3W0, Q9CRT7, Q9D0D7, Q9D116, Q9D4W4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf78 homolog. | |||||
|
SMC1A_HUMAN
|
||||||
| NC score | 0.064588 (rank : 23) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1346 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q14683, O14995, Q16351 | Gene names | SMC1L1, DXS423E, KIAA0178, SB1.8, SMC1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Sb1.8). | |||||
|
IF3A_HUMAN
|
||||||
| NC score | 0.064315 (rank : 24) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 875 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q14152, O00653 | Gene names | EIF3S10, KIAA0139 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a). | |||||
|
CEP70_HUMAN
|
||||||
| NC score | 0.064250 (rank : 25) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q8NHQ1, Q96B31, Q9H2C3, Q9H2Z1, Q9H940 | Gene names | CEP70, BITE | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 70 kDa (Cep70 protein) (p10-binding protein). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.063224 (rank : 26) | θ value | 0.62314 (rank : 15) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
IF3A_MOUSE
|
||||||
| NC score | 0.063137 (rank : 27) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 871 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P23116, Q60697, Q62162 | Gene names | Eif3s10, Csma, Eif3 | |||
|
Domain Architecture |

|
|||||
| Description | Eukaryotic translation initiation factor 3 subunit 10 (eIF-3 theta) (eIF3 p167) (eIF3 p180) (eIF3 p185) (eIF3a) (p162 protein) (Centrosomin). | |||||
|
SMC1A_MOUSE
|
||||||
| NC score | 0.062906 (rank : 28) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1384 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9CU62, Q3V480, Q9CUX9, Q9D959, Q9WTU1 | Gene names | Smc1l1, Sb1.8, Smc1, Smcb | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 1 protein (SMC1alpha protein) (Chromosome segregation protein SmcB) (Sb1.8). | |||||
|
CING_HUMAN
|
||||||
| NC score | 0.062249 (rank : 29) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1275 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q9P2M7, Q5T386, Q9NR25 | Gene names | CGN, KIAA1319 | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
TPR_HUMAN
|
||||||
| NC score | 0.062091 (rank : 30) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1921 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P12270, Q15655 | Gene names | TPR | |||
|
Domain Architecture |

|
|||||
| Description | Nucleoprotein TPR. | |||||
|
KTN1_MOUSE
|
||||||
| NC score | 0.061993 (rank : 31) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1324 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q61595, Q8BG49, Q8BHF4, Q8BHM8, Q8C9Y5, Q8CG51, Q8CG52, Q8CG53, Q8CG54, Q8CG55, Q8CG56, Q8CG57, Q8CG58, Q8CG59, Q8CG60, Q8CG61, Q8CG62, Q8CG63 | Gene names | Ktn1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinectin. | |||||
|
LIPA3_MOUSE
|
||||||
| NC score | 0.061861 (rank : 32) | θ value | 4.03905 (rank : 46) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P60469 | Gene names | Ppfia3 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-3 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-3) (PTPRF-interacting protein alpha-3). | |||||
|
CR034_MOUSE
|
||||||
| NC score | 0.061820 (rank : 33) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 643 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8CDV0 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C18orf34 homolog. | |||||
|
CI039_HUMAN
|
||||||
| NC score | 0.060619 (rank : 34) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1171 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9NXG0, Q5VYJ0, Q9HAJ5 | Gene names | C9orf39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C9orf39. | |||||
|
RAD50_MOUSE
|
||||||
| NC score | 0.060611 (rank : 35) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1366 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P70388, Q6PEB0, Q8C2T7, Q9CU59 | Gene names | Rad50 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | DNA repair protein RAD50 (EC 3.6.-.-) (mRad50). | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.060196 (rank : 36) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
LIPA1_HUMAN
|
||||||
| NC score | 0.060020 (rank : 37) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1294 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q13136, Q13135, Q14567, Q8N4I2 | Gene names | PPFIA1, LIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-alpha-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein alpha-1) (PTPRF-interacting protein alpha-1) (LAR-interacting protein 1) (LIP.1). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.059898 (rank : 38) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.059814 (rank : 39) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
LRRF2_HUMAN
|
||||||
| NC score | 0.059732 (rank : 40) | θ value | 8.99809 (rank : 67) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y608, Q68CV3, Q9NXH5 | Gene names | LRRFIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
EEA1_HUMAN
|
||||||
| NC score | 0.059720 (rank : 41) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1796 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q15075, Q14221 | Gene names | EEA1, ZFYVE2 | |||
|
Domain Architecture |

|
|||||
| Description | Early endosome antigen 1 (Endosome-associated protein p162) (Zinc finger FYVE domain-containing protein 2). | |||||
|
NUMA1_HUMAN
|
||||||
| NC score | 0.059374 (rank : 42) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1523 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q14980, Q14981 | Gene names | NUMA1, NUMA | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nuclear mitotic apparatus protein 1 (NuMA protein) (SP-H antigen). | |||||
|
CE152_HUMAN
|
||||||
| NC score | 0.059279 (rank : 43) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 976 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O94986, Q6NTA0 | Gene names | CEP152, KIAA0912 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 152 kDa (Cep152 protein). | |||||
|
CYTSA_MOUSE
|
||||||
| NC score | 0.058899 (rank : 44) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q2KN98, Q8CHF9 | Gene names | Cytsa, Kiaa0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A. | |||||
|
SM1L2_HUMAN
|
||||||
| NC score | 0.058847 (rank : 45) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1019 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8NDV3, Q5TIC3, Q9Y3G5 | Gene names | SMC1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 1-like 2 protein (SMC1beta protein). | |||||
|
CYTSA_HUMAN
|
||||||
| NC score | 0.058157 (rank : 46) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1138 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q69YQ0, O15081 | Gene names | CYTSA, KIAA0376 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytospin-A (NY-REN-22 antigen). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.058089 (rank : 47) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
CCD39_MOUSE
|
||||||
| NC score | 0.057929 (rank : 48) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1026 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9D5Y1, Q8CDG6 | Gene names | Ccdc39, D3Ertd789e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
NASP_HUMAN
|
||||||
| NC score | 0.057911 (rank : 49) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 709 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P49321, Q96A69, Q9BTW2 | Gene names | NASP | |||
|
Domain Architecture |

|
|||||
| Description | Nuclear autoantigenic sperm protein (NASP). | |||||
|
RB6I2_MOUSE
|
||||||
| NC score | 0.057802 (rank : 50) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q99MI1, Q80TK7, Q8BPL1, Q8C7Y1, Q99MI2 | Gene names | Erc1, Cast2, Kiaa1081, Rab6ip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1) (ERC1) (CAZ-associated structural protein 2) (CAST2). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.057758 (rank : 51) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
TRHY_HUMAN
|
||||||
| NC score | 0.057584 (rank : 52) | θ value | θ > 10 (rank : 194) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1385 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q07283 | Gene names | THH, THL, TRHY | |||
|
Domain Architecture |

|
|||||
| Description | Trichohyalin. | |||||
|
REST_HUMAN
|
||||||
| NC score | 0.057557 (rank : 53) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1605 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | P30622 | Gene names | RSN, CYLN1 | |||
|
Domain Architecture |

|
|||||
| Description | Restin (Cytoplasmic linker protein 170 alpha-2) (CLIP-170) (Reed- Sternberg intermediate filament-associated protein) (Cytoplasmic linker protein 1). | |||||
|
TRIPB_HUMAN
|
||||||
| NC score | 0.057512 (rank : 54) | θ value | θ > 10 (rank : 195) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1587 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15643, O14689, O15154, O95949 | Gene names | TRIP11, CEV14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thyroid receptor-interacting protein 11 (TRIP-11) (Golgi-associated microtubule-binding protein 210) (GMAP-210) (Trip230) (Clonal evolution-related gene on chromosome 14). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.057389 (rank : 55) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.057308 (rank : 56) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
RBCC1_HUMAN
|
||||||
| NC score | 0.057126 (rank : 57) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8TDY2, Q8WVU9, Q92601 | Gene names | RB1CC1, KIAA0203, RBICC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1. | |||||
|
PEPL_HUMAN
|
||||||
| NC score | 0.057083 (rank : 58) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1141 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O60437, O60314, O60454 | Gene names | PPL, KIAA0568 | |||
|
Domain Architecture |

|
|||||
| Description | Periplakin (195 kDa cornified envelope precursor protein) (190 kDa paraneoplastic pemphigus antigen). | |||||
|
EEA1_MOUSE
|
||||||
| NC score | 0.057041 (rank : 59) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1637 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8BL66, Q6DIC2 | Gene names | Eea1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Early endosome antigen 1. | |||||
|
APOA4_MOUSE
|
||||||
| NC score | 0.057017 (rank : 60) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P06728 | Gene names | Apoa4 | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein A-IV precursor (Apo-AIV) (ApoA-IV). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.056966 (rank : 61) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 65 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
SM1L2_MOUSE
|
||||||
| NC score | 0.056917 (rank : 62) | θ value | θ > 10 (rank : 181) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q920F6 | Gene names | Smc1l2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 1-like 2 protein (SMC1beta protein). | |||||
|
CP135_HUMAN
|
||||||
| NC score | 0.056665 (rank : 63) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1290 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q66GS9, O75130, Q9H8H7 | Gene names | CEP135, CEP4, KIAA0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
PIBF1_HUMAN
|
||||||
| NC score | 0.056547 (rank : 64) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8WXW3, O95664, Q6U9V2, Q6UG50, Q86V07, Q96SF4 | Gene names | PIBF1, C13orf24, PIBF | |||
|
Domain Architecture |

|
|||||
| Description | Progesterone-induced-blocking factor 1. | |||||
|
ERC2_MOUSE
|
||||||
| NC score | 0.056477 (rank : 65) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1166 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q6PH08, Q80U20, Q8CCP1 | Gene names | Erc2, Cast1, D14Ertd171e, Kiaa0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2 (CAZ-associated structural protein 1) (CAST1). | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.056472 (rank : 66) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
GOGA4_MOUSE
|
||||||
| NC score | 0.056414 (rank : 67) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1939 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q91VW5, O70365, Q8CGH6 | Gene names | Golga4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (tGolgin-1). | |||||
|
ERC2_HUMAN
|
||||||
| NC score | 0.056400 (rank : 68) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1156 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O15083, Q86TK4 | Gene names | ERC2, KIAA0378 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ERC protein 2. | |||||
|
GAS8_HUMAN
|
||||||
| NC score | 0.056272 (rank : 69) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 819 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O95995 | Gene names | GAS8, GAS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
KINH_MOUSE
|
||||||
| NC score | 0.056204 (rank : 70) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q61768, O08711, Q61580 | Gene names | Kif5b, Khcs, Kns1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
TMF1_HUMAN
|
||||||
| NC score | 0.056165 (rank : 71) | θ value | θ > 10 (rank : 192) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 987 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | P82094 | Gene names | TMF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | TATA element modulatory factor (TMF). | |||||
|
IFT74_HUMAN
|
||||||
| NC score | 0.056138 (rank : 72) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96LB3, Q6PGQ8, Q9H643, Q9H8G7 | Gene names | IFT74, CCDC2, CMG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
BPAEA_MOUSE
|
||||||
| NC score | 0.056049 (rank : 73) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1480 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | Q91ZU8, Q8K5D4 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoform 5 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
CI093_HUMAN
|
||||||
| NC score | 0.056006 (rank : 74) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1088 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q6TFL3, Q5SU58, Q6P1W1, Q6ZR13, Q8N1Z4, Q8N8L3, Q8TBR2 | Gene names | C9orf93 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C9orf93. | |||||
|
HMMR_MOUSE
|
||||||
| NC score | 0.055882 (rank : 75) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q00547 | Gene names | Hmmr, Ihabp, Rhamm | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility). | |||||
|
NIN_HUMAN
|
||||||
| NC score | 0.055828 (rank : 76) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1416 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8N4C6, Q6P0P6, Q9BWU6, Q9C012, Q9C013, Q9C014, Q9H5I6, Q9HAT7, Q9HBY5, Q9HCK7, Q9UH61 | Gene names | NIN, KIAA1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein (hNinein) (Glycogen synthase kinase 3 beta-interacting protein) (GSK3B-interacting protein). | |||||
|
GRAP1_HUMAN
|
||||||
| NC score | 0.055773 (rank : 77) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 944 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q4V328, Q4V327, Q4V330, Q5HYG1, Q6N046, Q96DH8, Q9NQ43, Q9ULQ3 | Gene names | GRIPAP1, KIAA1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1). | |||||
|
ZN365_MOUSE
|
||||||
| NC score | 0.055695 (rank : 78) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q8BG89, Q80TQ4, Q8BK39, Q8BXT2 | Gene names | Znf365, Kiaa0844, Zfp365 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein ZNF365 (Su48). | |||||
|
AKAP9_HUMAN
|
||||||
| NC score | 0.055132 (rank : 79) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1710 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q99996, O14869, O43355, O94895, Q9UQH3, Q9UQQ4, Q9Y6B8, Q9Y6Y2 | Gene names | AKAP9, AKAP350, AKAP450, KIAA0803 | |||
|
Domain Architecture |

|
|||||
| Description | A-kinase anchor protein 9 (Protein kinase A-anchoring protein 9) (PRKA9) (A-kinase anchor protein 450 kDa) (AKAP 450) (A-kinase anchor protein 350 kDa) (AKAP 350) (hgAKAP 350) (AKAP 120-like protein) (Protein hyperion) (Protein yotiao) (Centrosome- and Golgi-localized PKN-associated protein) (CG-NAP). | |||||
|
KIF5C_HUMAN
|
||||||
| NC score | 0.054960 (rank : 80) | θ value | 0.62314 (rank : 16) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 974 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | O60282, O95079 | Gene names | KIF5C, KIAA0531, NKHC2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.054904 (rank : 81) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
CCD46_HUMAN
|
||||||
| NC score | 0.054861 (rank : 82) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1430 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8N8E3, Q6PIB5, Q8NCR4, Q8NFR4 | Gene names | CCDC46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 46. | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.054857 (rank : 83) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.054851 (rank : 84) | θ value | 0.163984 (rank : 8) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
CEP63_HUMAN
|
||||||
| NC score | 0.054841 (rank : 85) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1005 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96MT8, Q96CR0, Q9H8F5, Q9H8N0 | Gene names | CEP63 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 63 kDa (Cep63 protein). | |||||
|
IFT74_MOUSE
|
||||||
| NC score | 0.054789 (rank : 86) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 939 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8BKE9, Q80ZI3, Q9CSY1, Q9CUS0, Q9D9T5 | Gene names | Ift74, Ccdc2, Cmg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 74 homolog (Coiled-coil domain-containing protein 2) (Capillary morphogenesis protein 1) (CMG-1). | |||||
|
GOGA3_MOUSE
|
||||||
| NC score | 0.054723 (rank : 87) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1437 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | P55937, Q80VF5, Q8CCK4, Q9QYT2, Q9QYT3 | Gene names | Golga3, Mea2 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Male-enhanced antigen 2) (MEA-2). | |||||
|
TAXB1_MOUSE
|
||||||
| NC score | 0.054707 (rank : 88) | θ value | θ > 10 (rank : 190) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 886 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q3UKC1, Q91YT6, Q9CVF0, Q9DC45 | Gene names | Tax1bp1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 homolog. | |||||
|
KNTC2_HUMAN
|
||||||
| NC score | 0.054651 (rank : 89) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 673 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | O14777, Q6PJX2 | Gene names | KNTC2, HEC, HEC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (HsHec1) (Kinetochore-associated protein 2) (Highly expressed in cancer protein) (Retinoblastoma-associated protein HEC). | |||||
|
NUF2_HUMAN
|
||||||
| NC score | 0.054608 (rank : 90) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 661 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9BZD4, Q8WU69, Q96HJ4, Q96Q78 | Gene names | CDCA1, NUF2, NUF2R | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (hNuf2) (hNuf2R) (hsNuf2) (Cell division cycle-associated protein 1). | |||||
|
CA065_HUMAN
|
||||||
| NC score | 0.054598 (rank : 91) | θ value | 3.0926 (rank : 36) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 469 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q8N715, Q8N746, Q8NA93 | Gene names | C1orf65 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C1orf65. | |||||
|
GOGA3_HUMAN
|
||||||
| NC score | 0.054573 (rank : 92) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1317 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q08378, O43241, Q6P9C7, Q86XW3, Q8TDA9, Q8WZA3 | Gene names | GOLGA3 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 3 (Golgin-160) (Golgi complex-associated protein of 170 kDa) (GCP170). | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.054529 (rank : 93) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
C102B_HUMAN
|
||||||
| NC score | 0.054510 (rank : 94) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 542 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q68D86, Q7Z467, Q8NDK7, Q9H5C1 | Gene names | CCDC102B, C18orf14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 102B. | |||||
|
GOGA5_HUMAN
|
||||||
| NC score | 0.054475 (rank : 95) | θ value | 8.99809 (rank : 65) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1168 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8TBA6, O95287, Q03962, Q9UQQ7 | Gene names | GOLGA5, RETII, RFG5 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (RET-fused gene 5 protein) (Ret-II protein). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.054395 (rank : 96) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
KNTC2_MOUSE
|
||||||
| NC score | 0.054270 (rank : 97) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 692 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q9D0F1, Q3TQT6, Q3UWM5, Q99P70 | Gene names | Kntc2, Hec1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Hec1 (Kinetochore-associated protein 2). | |||||
|
K0555_HUMAN
|
||||||
| NC score | 0.054250 (rank : 98) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 880 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q96AA8, O60302 | Gene names | KIAA0555 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0555. | |||||
|
HMMR_HUMAN
|
||||||
| NC score | 0.054239 (rank : 99) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1082 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | O75330, Q92767 | Gene names | HMMR, IHABP, RHAMM | |||
|
Domain Architecture |

|
|||||
| Description | Hyaluronan mediated motility receptor (Intracellular hyaluronic acid- binding protein) (Receptor for hyaluronan-mediated motility) (CD168 antigen). | |||||
|
KMHN1_HUMAN
|
||||||
| NC score | 0.054206 (rank : 100) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 950 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8TBZ0, Q86YI9, Q8N7W0 | Gene names | KMHN1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1. | |||||
|
GAS8_MOUSE
|
||||||
| NC score | 0.054163 (rank : 101) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 622 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q60779, Q99L71 | Gene names | Gas8, Gas11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Growth-arrest-specific protein 8 (Growth arrest-specific 11). | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.054140 (rank : 102) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
KMHN1_MOUSE
|
||||||
| NC score | 0.054008 (rank : 103) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 949 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q3V125, Q3TTQ8 | Gene names | Gm172 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cancer/testis antigen KM-HN-1 homolog. | |||||
|
RABE1_MOUSE
|
||||||
| NC score | 0.053862 (rank : 104) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1105 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O35551, Q99L08, Q9CRP3, Q9EQF9, Q9JI94 | Gene names | Rabep1, Rab5ep, Rabpt5, Rabpt5a | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha). | |||||
|
MD1L1_MOUSE
|
||||||
| NC score | 0.053861 (rank : 105) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1001 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9WTX8, Q9WTX9 | Gene names | Mad1l1, Mad1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1). | |||||
|
BICD1_MOUSE
|
||||||
| NC score | 0.053824 (rank : 106) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 837 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BR07, O55206, Q8BQ18, Q8BRR8, Q8C4D3, Q8CAK6, Q8R2J4, Q8R2J5, Q8R2J6, Q91YP7 | Gene names | Bicd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
CN145_MOUSE
|
||||||
| NC score | 0.053824 (rank : 107) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 916 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q8BI22 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145 homolog. | |||||
|
CJ118_MOUSE
|
||||||
| NC score | 0.053822 (rank : 108) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1132 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8C9S4, Q6P3D6, Q8CJC0 | Gene names | Otg1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 homolog (Oocyte-testis gene 1 protein). | |||||
|
MYH3_HUMAN
|
||||||
| NC score | 0.053764 (rank : 109) | θ value | 0.62314 (rank : 17) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1660 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P11055, Q15492 | Gene names | MYH3 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, fast skeletal muscle, embryonic (Muscle embryonic myosin heavy chain) (SMHCE). | |||||
|
SPC25_MOUSE
|
||||||
| NC score | 0.053694 (rank : 110) | θ value | θ > 10 (rank : 188) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 241 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q3UA16, Q9D021, Q9D1K6 | Gene names | Spbc25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Spc25. | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.053616 (rank : 111) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
NIN_MOUSE
|
||||||
| NC score | 0.053615 (rank : 112) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q61043, Q6ZPM7 | Gene names | Nin, Kiaa1565 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ninein. | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.053599 (rank : 113) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
CCD41_MOUSE
|
||||||
| NC score | 0.053430 (rank : 114) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1399 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9D5R3, Q3U7X7, Q3UX57, Q80VF0 | Gene names | Ccdc41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41. | |||||
|
CP250_HUMAN
|
||||||
| NC score | 0.053384 (rank : 115) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1805 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q9BV73, O14812, O60588, Q9H450 | Gene names | CEP250, CEP2, CNAP1 | |||
|
Domain Architecture |

|
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1). | |||||
|
SMC4_HUMAN
|
||||||
| NC score | 0.053314 (rank : 116) | θ value | θ > 10 (rank : 186) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9NTJ3, O95752, Q8NDL4, Q9UNT9 | Gene names | SMC4L1, CAPC, SMC4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (hCAP-C) (XCAP-C homolog). | |||||
|
HOOK1_HUMAN
|
||||||
| NC score | 0.053288 (rank : 117) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 942 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q9UJC3, O60561, Q5TG44 | Gene names | HOOK1 | |||
|
Domain Architecture |

|
|||||
| Description | Hook homolog 1 (h-hook1) (hHK1). | |||||
|
SMC2_MOUSE
|
||||||
| NC score | 0.053230 (rank : 118) | θ value | θ > 10 (rank : 183) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1131 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8CG48, Q61076, Q9CS17, Q9CSD8 | Gene names | Smc2l1, Cape, Fin16, Smc2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (XCAP-E homolog) (FGF-inducible protein 16). | |||||
|
CN145_HUMAN
|
||||||
| NC score | 0.053120 (rank : 119) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1196 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q6ZU80, Q96ML4 | Gene names | C14orf145 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C14orf145. | |||||
|
SAS6_MOUSE
|
||||||
| NC score | 0.053096 (rank : 120) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
TAXB1_HUMAN
|
||||||
| NC score | 0.053088 (rank : 121) | θ value | θ > 10 (rank : 189) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q86VP1, O60398, O95770, Q13311, Q9BQG5, Q9UI88 | Gene names | TAX1BP1, T6BP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tax1-binding protein 1 (TRAF6-binding protein). | |||||
|
CROCC_MOUSE
|
||||||
| NC score | 0.053049 (rank : 122) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1460 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8CJ40, Q7TQL2, Q80U01, Q8R0B9 | Gene names | Crocc, Kiaa0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
BICD1_HUMAN
|
||||||
| NC score | 0.053005 (rank : 123) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 844 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q96G01, O43892, O43893 | Gene names | BICD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 1. | |||||
|
CK5P2_MOUSE
|
||||||
| NC score | 0.052969 (rank : 124) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1151 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8K389, Q6PCN1, Q6ZPL0 | Gene names | Cdk5rap2, Kiaa1633 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48). | |||||
|
GRAP1_MOUSE
|
||||||
| NC score | 0.052954 (rank : 125) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1234 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8VD04, O35693, Q3T9C3, Q69ZP9 | Gene names | Gripap1, DXImx47e, Kiaa1167 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GRIP1-associated protein 1 (GRASP-1) (HCMV-interacting protein). | |||||
|
OPTN_HUMAN
|
||||||
| NC score | 0.052899 (rank : 126) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 827 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q96CV9, Q5T672, Q5T673, Q5T674, Q5T675, Q7LDL9, Q8N562, Q9UET9, Q9UEV4, Q9Y218 | Gene names | OPTN, FIP2, GLC1E, NRP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin (Optic neuropathy-inducing protein) (E3-14.7K-interacting protein) (FIP-2) (Huntingtin-interacting protein HYPL) (NEMO-related protein) (Transcription factor IIIA-interacting protein) (TFIIIA- IntP). | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.052796 (rank : 127) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
SAS6_HUMAN
|
||||||
| NC score | 0.052776 (rank : 128) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 791 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6UVJ0, Q8N3K0 | Gene names | SASS6, SAS6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog (HsSAS-6). | |||||
|
EVPL_HUMAN
|
||||||
| NC score | 0.052746 (rank : 129) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 895 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q92817 | Gene names | EVPL | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (210 kDa paraneoplastic pemphigus antigen) (p210) (210 kDa cornified envelope precursor protein). | |||||
|
TPM4_MOUSE
|
||||||
| NC score | 0.052735 (rank : 130) | θ value | 4.03905 (rank : 50) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 703 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q6IRU2 | Gene names | Tpm4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tropomyosin alpha-4 chain (Tropomyosin-4). | |||||
|
SMC4_MOUSE
|
||||||
| NC score | 0.052733 (rank : 131) | θ value | θ > 10 (rank : 187) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 830 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q8CG47, Q8BTS7, Q8BTY9, Q99K21 | Gene names | Smc4l1, Capc, Smc4 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosomes 4-like 1 protein (Chromosome- associated polypeptide C) (XCAP-C homolog). | |||||
|
CJ118_HUMAN
|
||||||
| NC score | 0.052694 (rank : 132) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 937 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q7Z3E2, Q2M2V6, Q6NS91, Q7RTP1, Q8N117, Q8N3G3, Q8N6C2, Q9NWA3 | Gene names | C10orf118 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein C10orf118 (CTCL tumor antigen HD-CL-01/L14-2). | |||||
|
EVPL_MOUSE
|
||||||
| NC score | 0.052691 (rank : 133) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 952 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9D952 | Gene names | Evpl | |||
|
Domain Architecture |

|
|||||
| Description | Envoplakin (p210) (210 kDa cornified envelope precursor protein). | |||||
|
MYH6_HUMAN
|
||||||
| NC score | 0.052678 (rank : 134) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1426 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | P13533, Q13943, Q14906, Q14907 | Gene names | MYH6, MYHCA | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle alpha isoform (MyHC-alpha). | |||||
|
SMC2_HUMAN
|
||||||
| NC score | 0.052523 (rank : 135) | θ value | θ > 10 (rank : 182) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O95347, Q9P1P2 | Gene names | SMC2L1, CAPE, SMC2 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 2-like 1 protein (Chromosome- associated protein E) (hCAP-E) (XCAP-E homolog). | |||||
|
SDCG8_HUMAN
|
||||||
| NC score | 0.052500 (rank : 136) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q86SQ7, O60527, Q3ZCR6, Q8N5F2, Q9P0F1 | Gene names | SDCCAG8, CCCAP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serologically defined colon cancer antigen 8 (Centrosomal colon cancer autoantigen protein) (hCCCAP) (Antigen NY-CO-8). | |||||
|
CCD13_HUMAN
|
||||||
| NC score | 0.052466 (rank : 137) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 721 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q8IYE1 | Gene names | CCDC13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 13. | |||||
|
MYH1_MOUSE
|
||||||
| NC score | 0.052402 (rank : 138) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1461 | Shared Neighborhood Hits | 60 | |
| SwissProt Accessions | Q5SX40 | Gene names | Myh1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-1 (Myosin heavy chain, skeletal muscle, adult 1). | |||||
|
GOGA1_HUMAN
|
||||||
| NC score | 0.052372 (rank : 139) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1222 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q92805, Q8IYZ9 | Gene names | GOLGA1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
RABE1_HUMAN
|
||||||
| NC score | 0.052371 (rank : 140) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1104 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q15276, O95369, Q8IVX3 | Gene names | RABEP1, RABPT5, RABPT5A | |||
|
Domain Architecture |

|
|||||
| Description | Rab GTPase-binding effector protein 1 (Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4) (NY-REN-17 antigen). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.052304 (rank : 141) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 61 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
CEP70_MOUSE
|
||||||
| NC score | 0.052244 (rank : 142) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 532 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | Q6IQY5, Q9CRL9, Q9CTS4, Q9JIC1 | Gene names | Cep70 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 70 kDa (Cep70 protein). | |||||
|
GOGA5_MOUSE
|
||||||
| NC score | 0.052239 (rank : 143) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1434 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9QYE6, O88317, Q3TGE7, Q3U6S5, Q3UUF9 | Gene names | Golga5, Retii | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 5 (Golgin-84) (Sumiko protein) (Ret-II protein). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.052217 (rank : 144) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.052212 (rank : 145) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
MYH8_MOUSE
|
||||||
| NC score | 0.052182 (rank : 146) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1600 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P13542, Q5SX36 | Gene names | Myh8, Myhsp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
CALD1_HUMAN
|
||||||
| NC score | 0.052165 (rank : 147) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q05682, Q13978, Q13979, Q14741, Q14742 | Gene names | CALD1, CAD, CDM | |||
|
Domain Architecture |

|
|||||
| Description | Caldesmon (CDM). | |||||
|
CCD41_HUMAN
|
||||||
| NC score | 0.052165 (rank : 148) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1139 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y592 | Gene names | CCDC41 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 41 (NY-REN-58 antigen). | |||||
|
SMC3_MOUSE
|
||||||
| NC score | 0.052162 (rank : 149) | θ value | θ > 10 (rank : 185) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1187 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9CW03, O35667, Q9QUS3 | Gene names | Cspg6, Bam, Bmh, Mmip1, Smc3, Smc3l1, Smcd | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome segregation protein SmcD) (Bamacan) (Basement membrane-associated chondroitin proteoglycan) (Mad member- interacting protein 1). | |||||
|
OPTN_MOUSE
|
||||||
| NC score | 0.051891 (rank : 150) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1120 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8K3K8 | Gene names | Optn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Optineurin. | |||||
|
KIF5C_MOUSE
|
||||||
| NC score | 0.051830 (rank : 151) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1040 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P28738, Q9Z2F8 | Gene names | Kif5c, Nkhc2 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain isoform 5C (Kinesin heavy chain neuron-specific 2). | |||||
|
LRRF2_MOUSE
|
||||||
| NC score | 0.051822 (rank : 152) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q91WK0, Q8BVD1, Q9CU89 | Gene names | Lrrfip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
SMC3_HUMAN
|
||||||
| NC score | 0.051722 (rank : 153) | θ value | θ > 10 (rank : 184) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1186 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9UQE7, O60464 | Gene names | CSPG6, BAM, BMH, SMC3, SMC3L1 | |||
|
Domain Architecture |

|
|||||
| Description | Structural maintenance of chromosome 3 (Chondroitin sulfate proteoglycan 6) (Chromosome-associated polypeptide) (hCAP) (Bamacan) (Basement membrane-associated chondroitin proteoglycan). | |||||
|
KIF5A_MOUSE
|
||||||
| NC score | 0.051667 (rank : 154) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 859 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P33175, Q5DTP1, Q6PDY7, Q9Z2F9 | Gene names | Kif5a, Kiaa4086, Kif5, Nkhc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinesin heavy chain isoform 5A (Neuronal kinesin heavy chain) (NKHC) (Kinesin heavy chain neuron-specific 1). | |||||
|
OFD1_HUMAN
|
||||||
| NC score | 0.051643 (rank : 155) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 753 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | O75665, O75666 | Gene names | OFD1, CXorf5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Oral-facial-digital syndrome 1 protein (Protein 71-7A). | |||||
|
CCD27_MOUSE
|
||||||
| NC score | 0.051535 (rank : 156) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q3V036 | Gene names | Ccdc27 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 27. | |||||
|
BICD2_MOUSE
|
||||||
| NC score | 0.051489 (rank : 157) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
GOGA6_HUMAN
|
||||||
| NC score | 0.051279 (rank : 158) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1007 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q9NYA3, Q9NYA7 | Gene names | GOLGA6, GLP | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 6 (Golgin linked to PML) (Golgin-like protein). | |||||
|
CCD19_HUMAN
|
||||||
| NC score | 0.051268 (rank : 159) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 639 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q9UL16 | Gene names | CCDC19, NESG1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 19 (Nasopharyngeal epithelium- specific protein 1). | |||||
|
INVO_HUMAN
|
||||||
| NC score | 0.051124 (rank : 160) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 636 | Shared Neighborhood Hits | 45 | |
| SwissProt Accessions | P07476 | Gene names | IVL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Involucrin. | |||||
|
CYLN2_HUMAN
|
||||||
| NC score | 0.051115 (rank : 161) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1004 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9UDT6, O14527, O43611 | Gene names | CYLN2, KIAA0291, WBSCR4, WSCR4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115) (Williams-Beuren syndrome chromosome region 4 protein). | |||||
|
CYLN2_MOUSE
|
||||||
| NC score | 0.051010 (rank : 162) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1002 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q9Z0H8, Q7TSI9, Q8CHU1, Q9EP81 | Gene names | Cyln2, Kiaa0291 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cytoplasmic linker protein 2 (Cytoplasmic linker protein 115) (CLIP- 115). | |||||
|
CEP55_MOUSE
|
||||||
| NC score | 0.050972 (rank : 163) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 693 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8BT07, Q8C2J0, Q8K2I8, Q8R2Y4, Q9DBZ8 | Gene names | Cep55 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome protein 55. | |||||
|
CK5P2_HUMAN
|
||||||
| NC score | 0.050944 (rank : 164) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1433 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q96SN8, Q7Z3L4, Q7Z3U1, Q7Z7I6, Q9BSW0, Q9H6J6, Q9HCD9, Q9NV90, Q9UIW9 | Gene names | CDK5RAP2, CEP215, KIAA1633 | |||
|
Domain Architecture |

|
|||||
| Description | CDK5 regulatory subunit-associated protein 2 (CDK5 activator-binding protein C48) (Centrosome-associated protein 215). | |||||
|
NUF2_MOUSE
|
||||||
| NC score | 0.050920 (rank : 165) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 585 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q99P69, Q8BTK7, Q8VE05, Q9CST5 | Gene names | Cdca1, Nuf2r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kinetochore protein Nuf2 (Cell division cycle-associated protein 1). | |||||
|
HAT1_HUMAN
|
||||||
| NC score | 0.050852 (rank : 166) | θ value | 4.03905 (rank : 44) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O14929, Q49A44, Q53QF0, Q53SU4 | Gene names | HAT1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Histone acetyltransferase type B catalytic subunit (EC 2.3.1.48). | |||||
|
CCD11_MOUSE
|
||||||
| NC score | 0.050737 (rank : 167) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 46 | |
| SwissProt Accessions | Q9D439 | Gene names | Ccdc11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 11. | |||||
|
UACA_HUMAN
|
||||||
| NC score | 0.050656 (rank : 168) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1622 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9BZF9, Q8N3B8, Q9HCL1, Q9NWC6 | Gene names | UACA, KIAA1561 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uveal autoantigen with coiled-coil domains and ankyrin repeats protein. | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.050650 (rank : 169) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
MYH7_HUMAN
|
||||||
| NC score | 0.050600 (rank : 170) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P12883, Q14836, Q14837, Q14904, Q16579, Q92679, Q9H1D5, Q9UMM8 | Gene names | MYH7, MYHCB | |||
|
Domain Architecture |

|
|||||
| Description | Myosin heavy chain, cardiac muscle beta isoform (MyHC-beta). | |||||
|
MD1L1_HUMAN
|
||||||
| NC score | 0.050520 (rank : 171) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 893 | Shared Neighborhood Hits | 55 | |
| SwissProt Accessions | Q9Y6D9, Q13312, Q75MI0, Q86UM4, Q9UNH0 | Gene names | MAD1L1, MAD1, TXBP181 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitotic spindle assembly checkpoint protein MAD1 (Mitotic arrest deficient-like protein 1) (MAD1-like 1) (Mitotic checkpoint MAD1 protein-homolog) (HsMAD1) (hMAD1) (Tax-binding protein 181). | |||||
|
3BP5_MOUSE
|
||||||
| NC score | 0.050462 (rank : 172) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q9Z131, Q8C903, Q8VCZ0 | Gene names | Sh3bp5, Sab | |||
|
Domain Architecture |

|
|||||
| Description | SH3 domain-binding protein 5 (SH3 domain-binding protein that preferentially associates with BTK). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.050297 (rank : 173) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
CCHCR_MOUSE
|
||||||
| NC score | 0.050231 (rank : 174) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 932 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | Q8K2I2 | Gene names | Cchcr1, Hcr | |||
|
Domain Architecture |

|
|||||
| Description | Coiled-coil alpha-helical rod protein 1 (Alpha helical coiled-coil rod protein). | |||||
|
LIPB1_MOUSE
|
||||||
| NC score | 0.050223 (rank : 175) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 727 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8C8U0, Q69ZN5, Q6GQV3, Q80VB4, Q9CUT7 | Gene names | Ppfibp1, Kiaa1230 | |||
|
Domain Architecture |

|
|||||
| Description | Liprin-beta-1 (Protein tyrosine phosphatase receptor type f polypeptide-interacting protein-binding protein 1) (PTPRF-interacting protein-binding protein 1). | |||||
|
TEX9_HUMAN
|
||||||
| NC score | 0.050064 (rank : 176) | θ value | θ > 10 (rank : 191) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 541 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8N6V9 | Gene names | TEX9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Testis-expressed sequence 9 protein. | |||||
|
NEBL_HUMAN
|
||||||
| NC score | 0.048217 (rank : 177) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | O76041, Q9UIC4 | Gene names | NEBL | |||
|
Domain Architecture |

|
|||||
| Description | Nebulette (Actin-binding Z-disk protein). | |||||
|
MY18B_HUMAN
|
||||||
| NC score | 0.047635 (rank : 178) | θ value | 8.99809 (rank : 68) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1299 | Shared Neighborhood Hits | 56 | |
| SwissProt Accessions | Q8IUG5, Q8NDI8, Q8TE65, Q8WWS0, Q96KH2, Q96KR8, Q96KR9 | Gene names | MYO18B | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18B (Myosin XVIIIb). | |||||
|
ANGL3_HUMAN
|
||||||
| NC score | 0.039471 (rank : 179) | θ value | 0.279714 (rank : 9) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9Y5C1 | Gene names | ANGPTL3, ANGPT5 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-related protein 3 precursor (Angiopoietin-like 3) (Angiopoietin-5) (ANG-5). | |||||
|
ANGP2_HUMAN
|
||||||
| NC score | 0.033906 (rank : 180) | θ value | 1.81305 (rank : 26) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 400 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | O15123, Q9NRR7, Q9P2Y7 | Gene names | ANGPT2 | |||
|
Domain Architecture |

|
|||||
| Description | Angiopoietin-2 precursor (ANG-2). | |||||
|
LAMB1_MOUSE
|
||||||
| NC score | 0.032522 (rank : 181) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 919 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P02469 | Gene names | Lamb1-1, Lamb-1 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-1 chain precursor (Laminin B1 chain). | |||||
|
ROCK1_HUMAN
|
||||||
| NC score | 0.031379 (rank : 182) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2000 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q13464 | Gene names | ROCK1 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 1 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 1) (p160 ROCK-1) (p160ROCK) (NY-REN-35 antigen). | |||||
|
LAMA4_MOUSE
|
||||||
| NC score | 0.029619 (rank : 183) | θ value | 8.99809 (rank : 66) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 584 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P97927, O88785, P70409 | Gene names | Lama4 | |||
|
Domain Architecture |

|
|||||
| Description | Laminin alpha-4 chain precursor. | |||||
|
MRCKB_HUMAN
|
||||||
| NC score | 0.028941 (rank : 184) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 2239 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q9Y5S2, Q2L7A5, Q86TJ1, Q9ULU5 | Gene names | CDC42BPB, KIAA1124 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK beta (EC 2.7.11.1) (CDC42-binding protein kinase beta) (Myotonic dystrophy kinase-related CDC42-binding kinase beta) (Myotonic dystrophy protein kinase-like beta) (MRCK beta) (DMPK-like beta). | |||||
|
DYH5_HUMAN
|
||||||
| NC score | 0.019033 (rank : 185) | θ value | 8.99809 (rank : 64) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 323 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q8TE73, Q92860, Q96L74, Q9H5S7, Q9HCG9 | Gene names | DNAH5, DNAHC5, KIAA1603 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1). | |||||
|
TAOK3_HUMAN
|
||||||
| NC score | 0.018234 (rank : 186) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1494 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q9H2K8, Q658N1, Q8IUM4, Q9HC79, Q9NZM9, Q9UHG7 | Gene names | TAOK3, DPK, JIK, KDS, MAP3K18 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3) (Jun kinase-inhibitory kinase) (JNK/SAPK- inhibitory kinase) (Dendritic cell-derived protein kinase) (Cutaneous T-cell lymphoma tumor antigen HD-CL-09) (CTCL tumor antigen HD-CL-09) (Kinase from chicken homolog A) (hKFC-A). | |||||
|
CETP_HUMAN
|
||||||
| NC score | 0.017833 (rank : 187) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P11597, Q13987, Q13988 | Gene names | CETP | |||
|
Domain Architecture |

|
|||||
| Description | Cholesteryl ester transfer protein precursor (Lipid transfer protein I). | |||||
|
TAOK3_MOUSE
|
||||||
| NC score | 0.016516 (rank : 188) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 1458 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q8BYC6, Q3V3B3 | Gene names | Taok3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase TAO3 (EC 2.7.11.1) (Thousand and one amino acid protein 3). | |||||
|
CN108_MOUSE
|
||||||
| NC score | 0.013228 (rank : 189) | θ value | 8.99809 (rank : 63) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8BJ63, Q3TPT5, Q8BII2, Q8BJ31, Q8BJ58, Q8BY11, Q8R5B1 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Protein C14orf108 homolog. | |||||
|
B4GT6_MOUSE
|
||||||
| NC score | 0.013219 (rank : 190) | θ value | 3.0926 (rank : 35) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9WVK5 | Gene names | B4galt6 | |||
|
Domain Architecture |

|
|||||
| Description | Beta-1,4-galactosyltransferase 6 (EC 2.4.1.-) (Beta-1,4-GalTase 6) (Beta4Gal-T6) (b4Gal-T6) (UDP-galactose:beta-N-acetylglucosamine beta- 1,4-galactosyltransferase 6) (UDP-Gal:beta-GlcNAc beta-1,4- galactosyltransferase 6) [Includes: Lactosylceramide synthase (EC 2.4.1.-) (LacCer synthase) (UDP-Gal:glucosylceramide beta-1,4- galactosyltransferase)]. | |||||
|
DSPP_MOUSE
|
||||||
| NC score | 0.009768 (rank : 191) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 491 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P97399, O70567 | Gene names | Dspp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dentin sialophosphoprotein precursor (Dentin matrix protein 3) (DMP-3) [Contains: Dentin phosphoprotein (Dentin phosphophoryn) (DPP); Dentin sialoprotein (DSP)]. | |||||
|
TRIM6_MOUSE
|
||||||
| NC score | 0.008017 (rank : 192) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8BGE7, Q99PQ6 | Gene names | Trim6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 6. | |||||
|
PARD3_HUMAN
|
||||||
| NC score | 0.007799 (rank : 193) | θ value | 8.99809 (rank : 69) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8TEW0, Q8TEW1, Q8TEW2, Q8TEW3, Q96K28, Q96RM6, Q96RM7, Q9BY57, Q9BY58, Q9HC48, Q9NWL4, Q9NYE6 | Gene names | PARD3, PAR3, PAR3A | |||
|
Domain Architecture |

|
|||||
| Description | Partitioning-defective 3 homolog (PARD-3) (PAR-3) (Atypical PKC isotype-specific-interacting protein) (ASIP) (CTCL tumor antigen se2- 5) (PAR3-alpha). | |||||
|
FLII_HUMAN
|
||||||
| NC score | 0.004918 (rank : 194) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 318 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q13045 | Gene names | FLII, FLIL | |||
|
Domain Architecture |

|
|||||
| Description | Protein flightless-1 homolog. | |||||
|
CHD9_MOUSE
|
||||||
| NC score | 0.004524 (rank : 195) | θ value | 8.99809 (rank : 62) | |||
| Query Neighborhood Hits | 72 | Target Neighborhood Hits | 427 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8BYH8, Q7TMV5, Q8BJG8, Q8BZJ2, Q8CHG8 | Gene names | Chd9, Kiaa0308, Pric320 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Chromodomain-helicase-DNA-binding protein 9 (EC 3.6.1.-) (ATP- dependent helicase CHD9) (CHD-9) (Peroxisomal proliferator-activated receptor A-interacting complex 320 kDa protein) (PPAR-alpha- interacting complex protein 320 kDa). | |||||