Please be patient as the page loads
|
SATB2_MOUSE
|
||||||
| SwissProt Accessions | Q8VI24 | Gene names | Satb2, Kiaa1034 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB2 (Special AT-rich sequence-binding protein 2). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SATB1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.970054 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q01826 | Gene names | SATB1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB1 (Special AT-rich sequence-binding protein 1). | |||||
|
SATB1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.970383 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q60611 | Gene names | Satb1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB1 (Special AT-rich sequence-binding protein 1). | |||||
|
SATB2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.999919 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UPW6 | Gene names | SATB2, KIAA1034 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB2 (Special AT-rich sequence-binding protein 2). | |||||
|
SATB2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8VI24 | Gene names | Satb2, Kiaa1034 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB2 (Special AT-rich sequence-binding protein 2). | |||||
|
ATBF1_HUMAN
|
||||||
| θ value | 2.44474e-05 (rank : 5) | NC score | 0.109770 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ATBF1_MOUSE
|
||||||
| θ value | 0.00020696 (rank : 6) | NC score | 0.107095 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
HNF6_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 7) | NC score | 0.266294 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBC0, Q99744, Q9UMR6 | Gene names | ONECUT1, HNF6, HNF6A | |||
|
Domain Architecture |
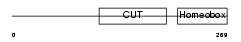
|
|||||
| Description | Hepatocyte nuclear factor 6 (HNF-6) (One cut domain family member 1). | |||||
|
HNF6_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 8) | NC score | 0.266485 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08755 | Gene names | Onecut1, Hnf6, Hnf6a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 6 (HNF-6) (One cut domain family member 1). | |||||
|
ONEC2_HUMAN
|
||||||
| θ value | 0.00035302 (rank : 9) | NC score | 0.275017 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95948 | Gene names | ONECUT2 | |||
|
Domain Architecture |

|
|||||
| Description | One cut domain family member 2 (Transcription factor ONECUT-2) (OC-2). | |||||
|
ONEC2_MOUSE
|
||||||
| θ value | 0.00035302 (rank : 10) | NC score | 0.273267 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6XBJ3 | Gene names | Onecut2, Oc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | One cut domain family member 2 (Transcription factor ONECUT-2) (OC-2). | |||||
|
CUTL2_MOUSE
|
||||||
| θ value | 0.0113563 (rank : 11) | NC score | 0.122261 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
CUTL2_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 12) | NC score | 0.118533 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
VSX1_MOUSE
|
||||||
| θ value | 0.0330416 (rank : 13) | NC score | 0.071864 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91V10 | Gene names | Vsx1, Rinx | |||
|
Domain Architecture |

|
|||||
| Description | Visual system homeobox 1 (Transcription factor VSX1) (Retinal inner nuclear layer homeobox protein) (Homeodomain protein RINX). | |||||
|
ESX1L_HUMAN
|
||||||
| θ value | 0.125558 (rank : 14) | NC score | 0.067807 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
SRPK1_HUMAN
|
||||||
| θ value | 0.163984 (rank : 15) | NC score | 0.025340 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96SB4, Q12890, Q5R364, Q5R365, Q8IY12 | Gene names | SRPK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
SHOX2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 16) | NC score | 0.069281 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60902, O60465, O60467, O60903 | Gene names | SHOX2, OG12X, SHOT | |||
|
Domain Architecture |

|
|||||
| Description | Short stature homeobox protein 2 (Paired-related homeobox protein SHOT) (Homeobox protein Og12X). | |||||
|
HOMEZ_MOUSE
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.126104 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80W88 | Gene names | Homez, Kiaa1443 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox and leucine zipper protein Homez (Homeodomain leucine zipper- containing factor). | |||||
|
SHOX2_MOUSE
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.066889 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70390, P70369 | Gene names | Shox2, Og12x, Prx3 | |||
|
Domain Architecture |

|
|||||
| Description | Short stature homeobox protein 2 (Homeobox protein Og12X) (OG-12) (Paired family homeodomain protein Prx3). | |||||
|
ZFHX2_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.088647 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
CUTL1_HUMAN
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.072170 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
ETV3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 21) | NC score | 0.025394 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
LHX61_HUMAN
|
||||||
| θ value | 1.38821 (rank : 22) | NC score | 0.062677 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UPM6, Q5T7S8, Q9NTK3, Q9UPM5 | Gene names | LHX6, LHX6.1 | |||
|
Domain Architecture |
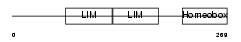
|
|||||
| Description | LIM/homeobox protein Lhx6.1 (Lhx6). | |||||
|
LHX6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 23) | NC score | 0.064673 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88706 | Gene names | Lhx6 | |||
|
Domain Architecture |
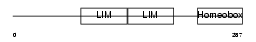
|
|||||
| Description | LIM/homeobox protein Lhx6. | |||||
|
PAX6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 24) | NC score | 0.054185 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P26367, Q6N006, Q99413 | Gene names | PAX6, AN2 | |||
|
Domain Architecture |
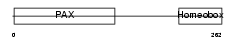
|
|||||
| Description | Paired box protein Pax-6 (Oculorhombin) (Aniridia type II protein). | |||||
|
PAX6_MOUSE
|
||||||
| θ value | 1.38821 (rank : 25) | NC score | 0.054185 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P63015, P32117, P70601, Q62222, Q64037, Q8CEI5, Q8VDB5, Q921Q8 | Gene names | Pax6, Pax-6, Sey | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paired box protein Pax-6 (Oculorhombin). | |||||
|
SHOX_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.059832 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15266, O00412, O00413, O15267 | Gene names | SHOX, PHOG | |||
|
Domain Architecture |

|
|||||
| Description | Short stature homeobox protein (Short stature homeobox-containing protein) (Pseudoautosomal homeobox-containing osteogenic protein). | |||||
|
ZHX2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.059128 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
RAI14_HUMAN
|
||||||
| θ value | 1.81305 (rank : 28) | NC score | 0.013784 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
RAI14_MOUSE
|
||||||
| θ value | 1.81305 (rank : 29) | NC score | 0.012691 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
TOX_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.036578 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
ALX4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 31) | NC score | 0.046341 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35137 | Gene names | Alx4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein aristaless-like 4 (ALX-4). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 2.36792 (rank : 32) | NC score | 0.064231 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
PAX4_MOUSE
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.046488 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P32115, P97341, Q9QUR1, Q9R091 | Gene names | Pax4, Pax-4 | |||
|
Domain Architecture |
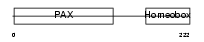
|
|||||
| Description | Paired box protein Pax-4. | |||||
|
SHOC2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.013107 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQ13, O76063 | Gene names | SHOC2, KIAA0862 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat protein SHOC-2 (Ras-binding protein Sur-8). | |||||
|
SHOC2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.013054 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88520, Q3UJH6, Q8BVL0 | Gene names | Shoc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat protein SHOC-2 (Ras-binding protein Sur-8). | |||||
|
TOX_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.034662 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.006856 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CSP9_HUMAN
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.049311 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43513 | Gene names | CRSP9, ARC34, MED7 | |||
|
Domain Architecture |
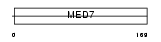
|
|||||
| Description | Cofactor required for Sp1 transcriptional activation subunit 9 (Transcriptional coactivator CRSP33) (RNA polymerase transcriptional regulation mediator subunit 7 homolog) (hMED7) (Activator-recruited cofactor 34 kDa component) (ARC34). | |||||
|
CSP9_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.049355 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CZB6, Q3UGV1, Q91VN3, Q9CWV8, Q9D0V9 | Gene names | Crsp9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cofactor required for Sp1 transcriptional activation subunit 9. | |||||
|
ATF6B_HUMAN
|
||||||
| θ value | 4.03905 (rank : 40) | NC score | 0.030978 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99941, Q13269, Q14343, Q14345, Q99635, Q99637, Q9H3V9, Q9H3W1, Q9NPL0 | Gene names | CREBL1, G13 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp) (Protein G13). | |||||
|
BRD4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 41) | NC score | 0.013198 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
DPP8_MOUSE
|
||||||
| θ value | 4.03905 (rank : 42) | NC score | 0.015337 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80YA7, Q9D4G6 | Gene names | Dpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dipeptidyl peptidase 8 (EC 3.4.14.5) (Dipeptidyl peptidase VIII) (DP8). | |||||
|
LHX61_MOUSE
|
||||||
| θ value | 4.03905 (rank : 43) | NC score | 0.055781 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R1R0, Q9R1R1 | Gene names | Lhx6.1 | |||
|
Domain Architecture |
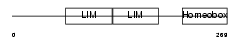
|
|||||
| Description | LIM/homeobox protein Lhx6.1. | |||||
|
HOMEZ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 44) | NC score | 0.097519 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IX15, Q6P049, Q86XB6, Q9P2A5 | Gene names | HOMEZ, KIAA1443 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox and leucine zipper protein Homez (Homeodomain leucine zipper- containing factor). | |||||
|
GSH2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 45) | NC score | 0.022193 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BZM3 | Gene names | GSH2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein GSH-2. | |||||
|
M3K4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 46) | NC score | 0.000750 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08648, O08649, O70124 | Gene names | Map3k4, Mekk4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4). | |||||
|
PO4F2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 47) | NC score | 0.030193 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q12837, Q13883, Q14987 | Gene names | POU4F2, BRN3B | |||
|
Domain Architecture |
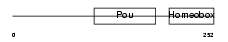
|
|||||
| Description | POU domain, class 4, transcription factor 2 (Brain-specific homeobox/POU domain protein 3B) (Brn-3B). | |||||
|
PO4F2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 48) | NC score | 0.030248 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q63934, Q63954 | Gene names | Pou4f2, Brn3b | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 4, transcription factor 2 (Brain-specific homeobox/POU domain protein 3B) (Brn-3B) (Brn-3.2). | |||||
|
SAMH1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 49) | NC score | 0.037023 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60710, Q91VK8 | Gene names | Samhd1, Mg11 | |||
|
Domain Architecture |
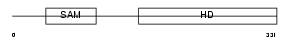
|
|||||
| Description | SAM domain and HD domain-containing protein 1 (Interferon-gamma- inducible protein Mg11). | |||||
|
SNIP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 50) | NC score | 0.019599 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
SRRM1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 51) | NC score | 0.011085 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
VSX1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 52) | NC score | 0.048324 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZR4, Q5TF40, Q5TF41, Q9HCU3, Q9NU27 | Gene names | VSX1, RINX | |||
|
Domain Architecture |

|
|||||
| Description | Visual system homeobox 1 (Transcription factor VSX1) (Retinal inner nuclear layer homeobox protein) (Homeodomain protein RINX). | |||||
|
BMP7_HUMAN
|
||||||
| θ value | 8.99809 (rank : 53) | NC score | 0.005447 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18075, Q9H512, Q9NTQ7 | Gene names | BMP7, OP1 | |||
|
Domain Architecture |
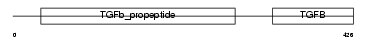
|
|||||
| Description | Bone morphogenetic protein 7 precursor (BMP-7) (Osteogenic protein 1) (OP-1) (Eptotermin alfa). | |||||
|
FUCO_MOUSE
|
||||||
| θ value | 8.99809 (rank : 54) | NC score | 0.013075 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99LJ1, Q3UAH8, Q8BN13, Q9DD22 | Gene names | Fuca1, Fuca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tissue alpha-L-fucosidase precursor (EC 3.2.1.51) (Alpha-L-fucosidase I) (Alpha-L-fucoside fucohydrolase). | |||||
|
PSL1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 55) | NC score | 0.015475 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
RGS3_MOUSE
|
||||||
| θ value | 8.99809 (rank : 56) | NC score | 0.009055 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
TIF1A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 57) | NC score | 0.007523 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
WNK4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 58) | NC score | 0.001411 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 961 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80UE6, Q4VAC1, Q80XB5, Q80XN2, Q8R0N0, Q8R340 | Gene names | Wnk4, Prkwnk4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
SATB2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8VI24 | Gene names | Satb2, Kiaa1034 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB2 (Special AT-rich sequence-binding protein 2). | |||||
|
SATB2_HUMAN
|
||||||
| NC score | 0.999919 (rank : 2) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q9UPW6 | Gene names | SATB2, KIAA1034 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB2 (Special AT-rich sequence-binding protein 2). | |||||
|
SATB1_MOUSE
|
||||||
| NC score | 0.970383 (rank : 3) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q60611 | Gene names | Satb1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB1 (Special AT-rich sequence-binding protein 1). | |||||
|
SATB1_HUMAN
|
||||||
| NC score | 0.970054 (rank : 4) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 130 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q01826 | Gene names | SATB1 | |||
|
Domain Architecture |

|
|||||
| Description | DNA-binding protein SATB1 (Special AT-rich sequence-binding protein 1). | |||||
|
ONEC2_HUMAN
|
||||||
| NC score | 0.275017 (rank : 5) | θ value | 0.00035302 (rank : 9) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O95948 | Gene names | ONECUT2 | |||
|
Domain Architecture |

|
|||||
| Description | One cut domain family member 2 (Transcription factor ONECUT-2) (OC-2). | |||||
|
ONEC2_MOUSE
|
||||||
| NC score | 0.273267 (rank : 6) | θ value | 0.00035302 (rank : 10) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6XBJ3 | Gene names | Onecut2, Oc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | One cut domain family member 2 (Transcription factor ONECUT-2) (OC-2). | |||||
|
HNF6_MOUSE
|
||||||
| NC score | 0.266485 (rank : 7) | θ value | 0.00035302 (rank : 8) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08755 | Gene names | Onecut1, Hnf6, Hnf6a | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 6 (HNF-6) (One cut domain family member 1). | |||||
|
HNF6_HUMAN
|
||||||
| NC score | 0.266294 (rank : 8) | θ value | 0.00035302 (rank : 7) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 127 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9UBC0, Q99744, Q9UMR6 | Gene names | ONECUT1, HNF6, HNF6A | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte nuclear factor 6 (HNF-6) (One cut domain family member 1). | |||||
|
HOMEZ_MOUSE
|
||||||
| NC score | 0.126104 (rank : 9) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 61 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q80W88 | Gene names | Homez, Kiaa1443 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox and leucine zipper protein Homez (Homeodomain leucine zipper- containing factor). | |||||
|
CUTL2_MOUSE
|
||||||
| NC score | 0.122261 (rank : 10) | θ value | 0.0113563 (rank : 11) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 771 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | P70298 | Gene names | Cutl2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
CUTL2_HUMAN
|
||||||
| NC score | 0.118533 (rank : 11) | θ value | 0.0330416 (rank : 12) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 906 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | O14529 | Gene names | CUTL2, KIAA0293 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 2 (Homeobox protein Cux-2) (Cut-like 2). | |||||
|
ATBF1_HUMAN
|
||||||
| NC score | 0.109770 (rank : 12) | θ value | 2.44474e-05 (rank : 5) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1547 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q15911, O15101, Q13719 | Gene names | ATBF1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
ATBF1_MOUSE
|
||||||
| NC score | 0.107095 (rank : 13) | θ value | 0.00020696 (rank : 6) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1841 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q61329 | Gene names | Atbf1 | |||
|
Domain Architecture |

|
|||||
| Description | Alpha-fetoprotein enhancer-binding protein (AT motif-binding factor) (AT-binding transcription factor 1). | |||||
|
HOMEZ_HUMAN
|
||||||
| NC score | 0.097519 (rank : 14) | θ value | 5.27518 (rank : 44) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q8IX15, Q6P049, Q86XB6, Q9P2A5 | Gene names | HOMEZ, KIAA1443 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox and leucine zipper protein Homez (Homeodomain leucine zipper- containing factor). | |||||
|
ZFHX2_HUMAN
|
||||||
| NC score | 0.088647 (rank : 15) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1178 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q9C0A1 | Gene names | ZFHX2, KIAA1762 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger homeobox protein 2. | |||||
|
CUTL1_HUMAN
|
||||||
| NC score | 0.072170 (rank : 16) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1574 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P39880, Q6NYH4, Q9UEV5 | Gene names | CUTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP). | |||||
|
VSX1_MOUSE
|
||||||
| NC score | 0.071864 (rank : 17) | θ value | 0.0330416 (rank : 13) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 356 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q91V10 | Gene names | Vsx1, Rinx | |||
|
Domain Architecture |

|
|||||
| Description | Visual system homeobox 1 (Transcription factor VSX1) (Retinal inner nuclear layer homeobox protein) (Homeodomain protein RINX). | |||||
|
SHOX2_HUMAN
|
||||||
| NC score | 0.069281 (rank : 18) | θ value | 0.365318 (rank : 16) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 353 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O60902, O60465, O60467, O60903 | Gene names | SHOX2, OG12X, SHOT | |||
|
Domain Architecture |

|
|||||
| Description | Short stature homeobox protein 2 (Paired-related homeobox protein SHOT) (Homeobox protein Og12X). | |||||
|
ESX1L_HUMAN
|
||||||
| NC score | 0.067807 (rank : 19) | θ value | 0.125558 (rank : 14) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 498 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8N693, Q7Z6K7 | Gene names | ESX1L, ESX1R | |||
|
Domain Architecture |

|
|||||
| Description | Extraembryonic, spermatogenesis, homeobox 1-like protein. | |||||
|
SHOX2_MOUSE
|
||||||
| NC score | 0.066889 (rank : 20) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 348 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P70390, P70369 | Gene names | Shox2, Og12x, Prx3 | |||
|
Domain Architecture |

|
|||||
| Description | Short stature homeobox protein 2 (Homeobox protein Og12X) (OG-12) (Paired family homeodomain protein Prx3). | |||||
|
LHX6_MOUSE
|
||||||
| NC score | 0.064673 (rank : 21) | θ value | 1.38821 (rank : 23) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 399 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | O88706 | Gene names | Lhx6 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx6. | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.064231 (rank : 22) | θ value | 2.36792 (rank : 32) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
LHX61_HUMAN
|
||||||
| NC score | 0.062677 (rank : 23) | θ value | 1.38821 (rank : 22) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 412 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9UPM6, Q5T7S8, Q9NTK3, Q9UPM5 | Gene names | LHX6, LHX6.1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx6.1 (Lhx6). | |||||
|
SHOX_HUMAN
|
||||||
| NC score | 0.059832 (rank : 24) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | O15266, O00412, O00413, O15267 | Gene names | SHOX, PHOG | |||
|
Domain Architecture |

|
|||||
| Description | Short stature homeobox protein (Short stature homeobox-containing protein) (Pseudoautosomal homeobox-containing osteogenic protein). | |||||
|
ZHX2_MOUSE
|
||||||
| NC score | 0.059128 (rank : 25) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q8C0C0 | Gene names | Zhx2, Afr1, Raf | |||
|
Domain Architecture |

|
|||||
| Description | Zinc fingers and homeoboxes protein 2 (Zinc finger and homeodomain protein 2) (Alpha-fetoprotein regulator 1) (AFP regulator 1) (Regulator of AFP). | |||||
|
LHX61_MOUSE
|
||||||
| NC score | 0.055781 (rank : 26) | θ value | 4.03905 (rank : 43) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 396 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q9R1R0, Q9R1R1 | Gene names | Lhx6.1 | |||
|
Domain Architecture |

|
|||||
| Description | LIM/homeobox protein Lhx6.1. | |||||
|
PAX6_HUMAN
|
||||||
| NC score | 0.054185 (rank : 27) | θ value | 1.38821 (rank : 24) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P26367, Q6N006, Q99413 | Gene names | PAX6, AN2 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-6 (Oculorhombin) (Aniridia type II protein). | |||||
|
PAX6_MOUSE
|
||||||
| NC score | 0.054185 (rank : 28) | θ value | 1.38821 (rank : 25) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 383 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P63015, P32117, P70601, Q62222, Q64037, Q8CEI5, Q8VDB5, Q921Q8 | Gene names | Pax6, Pax-6, Sey | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Paired box protein Pax-6 (Oculorhombin). | |||||
|
CSP9_MOUSE
|
||||||
| NC score | 0.049355 (rank : 29) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CZB6, Q3UGV1, Q91VN3, Q9CWV8, Q9D0V9 | Gene names | Crsp9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cofactor required for Sp1 transcriptional activation subunit 9. | |||||
|
CSP9_HUMAN
|
||||||
| NC score | 0.049311 (rank : 30) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43513 | Gene names | CRSP9, ARC34, MED7 | |||
|
Domain Architecture |

|
|||||
| Description | Cofactor required for Sp1 transcriptional activation subunit 9 (Transcriptional coactivator CRSP33) (RNA polymerase transcriptional regulation mediator subunit 7 homolog) (hMED7) (Activator-recruited cofactor 34 kDa component) (ARC34). | |||||
|
VSX1_HUMAN
|
||||||
| NC score | 0.048324 (rank : 31) | θ value | 6.88961 (rank : 52) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 344 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q9NZR4, Q5TF40, Q5TF41, Q9HCU3, Q9NU27 | Gene names | VSX1, RINX | |||
|
Domain Architecture |

|
|||||
| Description | Visual system homeobox 1 (Transcription factor VSX1) (Retinal inner nuclear layer homeobox protein) (Homeodomain protein RINX). | |||||
|
PAX4_MOUSE
|
||||||
| NC score | 0.046488 (rank : 32) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 370 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P32115, P97341, Q9QUR1, Q9R091 | Gene names | Pax4, Pax-4 | |||
|
Domain Architecture |

|
|||||
| Description | Paired box protein Pax-4. | |||||
|
ALX4_MOUSE
|
||||||
| NC score | 0.046341 (rank : 33) | θ value | 2.36792 (rank : 31) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | O35137 | Gene names | Alx4 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein aristaless-like 4 (ALX-4). | |||||
|
SAMH1_MOUSE
|
||||||
| NC score | 0.037023 (rank : 34) | θ value | 6.88961 (rank : 49) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q60710, Q91VK8 | Gene names | Samhd1, Mg11 | |||
|
Domain Architecture |

|
|||||
| Description | SAM domain and HD domain-containing protein 1 (Interferon-gamma- inducible protein Mg11). | |||||
|
TOX_MOUSE
|
||||||
| NC score | 0.036578 (rank : 35) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q66JW3, Q8BKH9, Q8BYQ5, Q8R4H0 | Gene names | Tox | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
TOX_HUMAN
|
||||||
| NC score | 0.034662 (rank : 36) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 106 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O94900, Q96AV5 | Gene names | TOX, KIAA0808 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Thymus high mobility group box protein TOX. | |||||
|
ATF6B_HUMAN
|
||||||
| NC score | 0.030978 (rank : 37) | θ value | 4.03905 (rank : 40) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 122 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99941, Q13269, Q14343, Q14345, Q99635, Q99637, Q9H3V9, Q9H3W1, Q9NPL0 | Gene names | CREBL1, G13 | |||
|
Domain Architecture |

|
|||||
| Description | Cyclic AMP-dependent transcription factor ATF-6 beta (Activating transcription factor 6 beta) (ATF6-beta) (cAMP-responsive element- binding protein-like 1) (cAMP response element-binding protein-related protein) (Creb-rp) (Protein G13). | |||||
|
PO4F2_MOUSE
|
||||||
| NC score | 0.030248 (rank : 38) | θ value | 6.88961 (rank : 48) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q63934, Q63954 | Gene names | Pou4f2, Brn3b | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 4, transcription factor 2 (Brain-specific homeobox/POU domain protein 3B) (Brn-3B) (Brn-3.2). | |||||
|
PO4F2_HUMAN
|
||||||
| NC score | 0.030193 (rank : 39) | θ value | 6.88961 (rank : 47) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q12837, Q13883, Q14987 | Gene names | POU4F2, BRN3B | |||
|
Domain Architecture |

|
|||||
| Description | POU domain, class 4, transcription factor 2 (Brain-specific homeobox/POU domain protein 3B) (Brn-3B). | |||||
|
ETV3_HUMAN
|
||||||
| NC score | 0.025394 (rank : 40) | θ value | 1.06291 (rank : 21) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 312 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P41162, Q8TAC8, Q9BX30 | Gene names | ETV3, METS, PE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ETS translocation variant 3 (ETS-domain transcriptional repressor PE1) (PE-1) (Mitogenic Ets transcriptional suppressor). | |||||
|
SRPK1_HUMAN
|
||||||
| NC score | 0.025340 (rank : 41) | θ value | 0.163984 (rank : 15) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 706 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96SB4, Q12890, Q5R364, Q5R365, Q8IY12 | Gene names | SRPK1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase SRPK1 (EC 2.7.11.1) (Serine/arginine- rich protein-specific kinase 1) (SR-protein-specific kinase 1) (SFRS protein kinase 1). | |||||
|
GSH2_HUMAN
|
||||||
| NC score | 0.022193 (rank : 42) | θ value | 6.88961 (rank : 45) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 23 | |
| SwissProt Accessions | Q9BZM3 | Gene names | GSH2 | |||
|
Domain Architecture |

|
|||||
| Description | Homeobox protein GSH-2. | |||||
|
SNIP_MOUSE
|
||||||
| NC score | 0.019599 (rank : 43) | θ value | 6.88961 (rank : 50) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 319 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9QWI6, O70298 | Gene names | P140, Kiaa1684 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | p130Cas-associated protein (p140Cap) (SNAP-25-interacting protein) (SNIP). | |||||
|
PSL1_HUMAN
|
||||||
| NC score | 0.015475 (rank : 44) | θ value | 8.99809 (rank : 55) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 650 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8TCT7, O60365, Q567S3, Q8IUH9, Q9BUY6, Q9H3M4, Q9NPN2, Q9P1Z6 | Gene names | SPPL2B, KIAA1532, PSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal peptide peptidase-like 2B (EC 3.4.23.-) (Protein SPP-like 2B) (Protein SPPL2b) (Intramembrane protease 4) (IMP4) (Presenilin-like protein 1). | |||||
|
DPP8_MOUSE
|
||||||
| NC score | 0.015337 (rank : 45) | θ value | 4.03905 (rank : 42) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q80YA7, Q9D4G6 | Gene names | Dpp8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dipeptidyl peptidase 8 (EC 3.4.14.5) (Dipeptidyl peptidase VIII) (DP8). | |||||
|
RAI14_HUMAN
|
||||||
| NC score | 0.013784 (rank : 46) | θ value | 1.81305 (rank : 28) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1367 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q9P0K7, Q6V1W9, Q7Z5I4, Q7Z733, Q9P2L2, Q9Y3T5 | Gene names | RAI14, KIAA1334, NORPEG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein). | |||||
|
BRD4_HUMAN
|
||||||
| NC score | 0.013198 (rank : 47) | θ value | 4.03905 (rank : 41) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 851 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | O60885, O60433, Q96PD3 | Gene names | BRD4, HUNK1 | |||
|
Domain Architecture |

|
|||||
| Description | Bromodomain-containing protein 4 (HUNK1 protein). | |||||
|
SHOC2_HUMAN
|
||||||
| NC score | 0.013107 (rank : 48) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UQ13, O76063 | Gene names | SHOC2, KIAA0862 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat protein SHOC-2 (Ras-binding protein Sur-8). | |||||
|
FUCO_MOUSE
|
||||||
| NC score | 0.013075 (rank : 49) | θ value | 8.99809 (rank : 54) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q99LJ1, Q3UAH8, Q8BN13, Q9DD22 | Gene names | Fuca1, Fuca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tissue alpha-L-fucosidase precursor (EC 3.2.1.51) (Alpha-L-fucosidase I) (Alpha-L-fucoside fucohydrolase). | |||||
|
SHOC2_MOUSE
|
||||||
| NC score | 0.013054 (rank : 50) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 308 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88520, Q3UJH6, Q8BVL0 | Gene names | Shoc2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat protein SHOC-2 (Ras-binding protein Sur-8). | |||||
|
RAI14_MOUSE
|
||||||
| NC score | 0.012691 (rank : 51) | θ value | 1.81305 (rank : 29) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1505 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9EP71, Q3URT3, Q6ZPT6 | Gene names | Rai14, Kiaa1334, Norpeg | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankycorbin (Ankyrin repeat and coiled-coil structure-containing protein) (Retinoic acid-induced protein 14) (Novel retinal pigment epithelial cell protein) (p125). | |||||
|
SRRM1_MOUSE
|
||||||
| NC score | 0.011085 (rank : 52) | θ value | 6.88961 (rank : 51) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 966 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q52KI8, O70495, Q9CVG5 | Gene names | Srrm1, Pop101 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/arginine repetitive matrix protein 1 (Plenty-of-prolines 101). | |||||
|
RGS3_MOUSE
|
||||||
| NC score | 0.009055 (rank : 53) | θ value | 8.99809 (rank : 56) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 1621 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q9DC04, Q5SRB1, Q5SRB4, Q5SRB8, Q8CEE3, Q8VI25, Q925G9, Q9JL22, Q9JL23, Q9QXA2 | Gene names | Rgs3 | |||
|
Domain Architecture |

|
|||||
| Description | Regulator of G-protein signaling 3 (RGS3) (C2PA). | |||||
|
TIF1A_MOUSE
|
||||||
| NC score | 0.007523 (rank : 54) | θ value | 8.99809 (rank : 57) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q64127, Q64126 | Gene names | Trim24, Tif1, Tif1a | |||
|
Domain Architecture |

|
|||||
| Description | Transcription intermediary factor 1-alpha (TIF1-alpha) (Tripartite motif-containing protein 24). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.006856 (rank : 55) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
BMP7_HUMAN
|
||||||
| NC score | 0.005447 (rank : 56) | θ value | 8.99809 (rank : 53) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P18075, Q9H512, Q9NTQ7 | Gene names | BMP7, OP1 | |||
|
Domain Architecture |

|
|||||
| Description | Bone morphogenetic protein 7 precursor (BMP-7) (Osteogenic protein 1) (OP-1) (Eptotermin alfa). | |||||
|
WNK4_MOUSE
|
||||||
| NC score | 0.001411 (rank : 57) | θ value | 8.99809 (rank : 58) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 961 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q80UE6, Q4VAC1, Q80XB5, Q80XN2, Q8R0N0, Q8R340 | Gene names | Wnk4, Prkwnk4 | |||
|
Domain Architecture |

|
|||||
| Description | Serine/threonine-protein kinase WNK4 (EC 2.7.11.1) (Protein kinase with no lysine 4) (Protein kinase, lysine-deficient 4). | |||||
|
M3K4_MOUSE
|
||||||
| NC score | 0.000750 (rank : 58) | θ value | 6.88961 (rank : 46) | |||
| Query Neighborhood Hits | 58 | Target Neighborhood Hits | 924 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O08648, O08649, O70124 | Gene names | Map3k4, Mekk4 | |||
|
Domain Architecture |

|
|||||
| Description | Mitogen-activated protein kinase kinase kinase 4 (EC 2.7.11.25) (MAPK/ERK kinase kinase 4) (MEK kinase 4) (MEKK 4). | |||||