Please be patient as the page loads
|
SAMD9_HUMAN
|
||||||
| SwissProt Accessions | Q5K651, Q6P080, Q75N21, Q8IVG6, Q9NXS8 | Gene names | SAMD9, KIAA2004, OEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 9. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
SAMD9_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q5K651, Q6P080, Q75N21, Q8IVG6, Q9NXS8 | Gene names | SAMD9, KIAA2004, OEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 9. | |||||
|
SHAN2_HUMAN
|
||||||
| θ value | 6.43352e-06 (rank : 2) | NC score | 0.221531 (rank : 2) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
SHAN2_MOUSE
|
||||||
| θ value | 6.43352e-06 (rank : 3) | NC score | 0.210563 (rank : 3) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80Z38, Q3UTK4, Q5DU07 | Gene names | Shank2, Kiaa1022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2) (Cortactin- binding protein 1) (CortBP1). | |||||
|
PHC1_MOUSE
|
||||||
| θ value | 0.00509761 (rank : 4) | NC score | 0.169301 (rank : 5) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64028, P70359, Q64307, Q8BZ80 | Gene names | Phc1, Edr, Edr1, Rae28 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (mPH1) (Early development regulatory protein 1) (RAE-28). | |||||
|
PHC1_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 5) | NC score | 0.172095 (rank : 4) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P78364, Q8WVM3, Q9BU63 | Gene names | PHC1, EDR1, PH1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (hPH1) (Early development regulatory protein 1). | |||||
|
PHC2_HUMAN
|
||||||
| θ value | 0.0193708 (rank : 6) | NC score | 0.153208 (rank : 7) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
PHC2_MOUSE
|
||||||
| θ value | 0.0193708 (rank : 7) | NC score | 0.163440 (rank : 6) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QWH1, O88463, Q8K5D9 | Gene names | Phc2, Edr2, Ph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (mPH2) (Early development regulatory protein 2) (p36). | |||||
|
SHAN1_HUMAN
|
||||||
| θ value | 0.0252991 (rank : 8) | NC score | 0.080451 (rank : 16) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
NOP56_MOUSE
|
||||||
| θ value | 0.163984 (rank : 9) | NC score | 0.089265 (rank : 12) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
GOGA1_MOUSE
|
||||||
| θ value | 0.21417 (rank : 10) | NC score | 0.038389 (rank : 31) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
MAP1B_MOUSE
|
||||||
| θ value | 0.21417 (rank : 11) | NC score | 0.054113 (rank : 23) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
NEB1_HUMAN
|
||||||
| θ value | 0.21417 (rank : 12) | NC score | 0.065129 (rank : 20) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
IFIT2_HUMAN
|
||||||
| θ value | 0.365318 (rank : 13) | NC score | 0.069732 (rank : 17) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09913 | Gene names | IFIT2, G10P2, IFI54 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 2 (IFIT-2) (Interferon-induced 54 kDa protein) (IFI-54K) (ISG-54 K). | |||||
|
IFIT2_MOUSE
|
||||||
| θ value | 0.47712 (rank : 14) | NC score | 0.069472 (rank : 18) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64112, Q62385 | Gene names | Ifit2, Garg39, Ifi54 | |||
|
Domain Architecture |
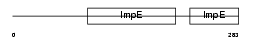
|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 2 (IFIT-2) (Interferon-induced 54 kDa protein) (IFI-54K) (Glucocorticoid- attenuated response gene 39 protein) (GARG-39). | |||||
|
MGAT5_MOUSE
|
||||||
| θ value | 0.47712 (rank : 15) | NC score | 0.084596 (rank : 13) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4G6 | Gene names | Mgat5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V (EC 2.4.1.155) (Mannoside acetylglucosaminyltransferase 5) (Alpha- mannoside beta-1,6-N-acetylglucosaminyltransferase) (N- acetylglucosaminyl-transferase V) (GNT-V) (GlcNAc-T V). | |||||
|
NOP56_HUMAN
|
||||||
| θ value | 0.47712 (rank : 16) | NC score | 0.081208 (rank : 15) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00567, Q9NQ05 | Gene names | NOL5A, NOP56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
PHC3_HUMAN
|
||||||
| θ value | 0.47712 (rank : 17) | NC score | 0.129003 (rank : 8) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
MGAT5_HUMAN
|
||||||
| θ value | 0.62314 (rank : 18) | NC score | 0.082682 (rank : 14) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q09328 | Gene names | MGAT5, GGNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V (EC 2.4.1.155) (Mannoside acetylglucosaminyltransferase 5) (Alpha- mannoside beta-1,6-N-acetylglucosaminyltransferase) (N- acetylglucosaminyl-transferase V) (GNT-V) (GlcNAc-T V). | |||||
|
MYO5A_HUMAN
|
||||||
| θ value | 0.62314 (rank : 19) | NC score | 0.031652 (rank : 36) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
MPP10_MOUSE
|
||||||
| θ value | 0.813845 (rank : 20) | NC score | 0.054389 (rank : 21) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
NFM_MOUSE
|
||||||
| θ value | 0.813845 (rank : 21) | NC score | 0.028398 (rank : 49) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
SAM14_MOUSE
|
||||||
| θ value | 0.813845 (rank : 22) | NC score | 0.114134 (rank : 11) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K070, Q5SWB7, Q8BHE2, Q8C8N5 | Gene names | Samd14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 14. | |||||
|
CE290_HUMAN
|
||||||
| θ value | 1.06291 (rank : 23) | NC score | 0.035642 (rank : 32) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
CF066_MOUSE
|
||||||
| θ value | 1.06291 (rank : 24) | NC score | 0.067567 (rank : 19) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D1H6, Q3TV72 | Gene names | ||||
|
Domain Architecture |
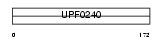
|
|||||
| Description | UPF0240 protein C6orf66 homolog. | |||||
|
SAM14_HUMAN
|
||||||
| θ value | 1.06291 (rank : 25) | NC score | 0.118797 (rank : 10) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZD0, Q8N2X0 | Gene names | SAMD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 14. | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 1.38821 (rank : 26) | NC score | 0.032441 (rank : 34) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MYO5A_MOUSE
|
||||||
| θ value | 1.38821 (rank : 27) | NC score | 0.029802 (rank : 43) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
PHC3_MOUSE
|
||||||
| θ value | 1.38821 (rank : 28) | NC score | 0.123773 (rank : 9) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
RBBP6_HUMAN
|
||||||
| θ value | 1.38821 (rank : 29) | NC score | 0.042845 (rank : 27) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
AFF4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 30) | NC score | 0.033765 (rank : 33) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
AP3B1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 31) | NC score | 0.027605 (rank : 51) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 1.81305 (rank : 32) | NC score | 0.028708 (rank : 45) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
ANKR5_MOUSE
|
||||||
| θ value | 2.36792 (rank : 33) | NC score | 0.016332 (rank : 68) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D2J7 | Gene names | Ankrd5 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 5. | |||||
|
DEK_MOUSE
|
||||||
| θ value | 2.36792 (rank : 34) | NC score | 0.046961 (rank : 26) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
GBP2_HUMAN
|
||||||
| θ value | 2.36792 (rank : 35) | NC score | 0.030780 (rank : 41) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P32456, Q86TB0 | Gene names | GBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 2 (GTP-binding protein 2) (Guanine nucleotide-binding protein 2) (GBP-2) (HuGBP-2). | |||||
|
IFIT3_HUMAN
|
||||||
| θ value | 2.36792 (rank : 36) | NC score | 0.054155 (rank : 22) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14879, Q99634, Q9BSK7 | Gene names | IFIT3, IFI60, IFIT4 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 3 (IFIT-3) (IFIT-4) (Interferon-induced 60 kDa protein) (IFI-60K) (ISG-60) (CIG49) (Retinoic acid-induced gene G protein) (RIG-G). | |||||
|
AREG_MOUSE
|
||||||
| θ value | 3.0926 (rank : 37) | NC score | 0.040051 (rank : 29) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31955 | Gene names | Areg, Sdgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amphiregulin precursor (AR) (Schwannoma-derived growth factor) (SDGF). | |||||
|
BRDT_MOUSE
|
||||||
| θ value | 3.0926 (rank : 38) | NC score | 0.019928 (rank : 65) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91Y44, Q59HJ4 | Gene names | Brdt, Fsrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein) (Bromodomain- containing female sterile homeotic-like protein). | |||||
|
MFN1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 39) | NC score | 0.025487 (rank : 57) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q811U4, Q811D5, Q8CEY6, Q99M10, Q9D395 | Gene names | Mfn1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN1 (EC 3.6.5.-) (Mitofusin-1). | |||||
|
NFM_HUMAN
|
||||||
| θ value | 3.0926 (rank : 40) | NC score | 0.026247 (rank : 53) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
RBCC1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 41) | NC score | 0.031319 (rank : 37) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
SFRIP_HUMAN
|
||||||
| θ value | 3.0926 (rank : 42) | NC score | 0.042013 (rank : 28) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
ZN148_HUMAN
|
||||||
| θ value | 3.0926 (rank : 43) | NC score | 0.008619 (rank : 71) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UQR1, O00389, O43591 | Gene names | ZNF148, ZBP89 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89). | |||||
|
ZN148_MOUSE
|
||||||
| θ value | 3.0926 (rank : 44) | NC score | 0.008708 (rank : 70) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61624, P97475 | Gene names | Znf148, Zbp89, Zfp148 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89) (G-rich box-binding protein) (Beta enolase repressor factor 1) (Transcription factor BFCOL1). | |||||
|
ZN291_HUMAN
|
||||||
| θ value | 3.0926 (rank : 45) | NC score | 0.039069 (rank : 30) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
ZRF1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 46) | NC score | 0.030841 (rank : 40) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
TCOF_HUMAN
|
||||||
| θ value | 4.03905 (rank : 47) | NC score | 0.031243 (rank : 39) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
TIAM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 48) | NC score | 0.022283 (rank : 62) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
ZN484_HUMAN
|
||||||
| θ value | 4.03905 (rank : 49) | NC score | 0.002532 (rank : 78) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 766 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5JVG2 | Gene names | ZNF484 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 484. | |||||
|
CS007_HUMAN
|
||||||
| θ value | 5.27518 (rank : 50) | NC score | 0.026056 (rank : 55) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CS007_MOUSE
|
||||||
| θ value | 5.27518 (rank : 51) | NC score | 0.027794 (rank : 50) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
GOGB1_HUMAN
|
||||||
| θ value | 5.27518 (rank : 52) | NC score | 0.028960 (rank : 44) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
TSH2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 53) | NC score | 0.014680 (rank : 69) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
ZN485_HUMAN
|
||||||
| θ value | 5.27518 (rank : 54) | NC score | 0.002362 (rank : 79) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NCK3, Q96CL0 | Gene names | ZNF485 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 485. | |||||
|
CC019_HUMAN
|
||||||
| θ value | 6.88961 (rank : 55) | NC score | 0.025734 (rank : 56) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PII3, Q96CS5 | Gene names | C3orf19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf19. | |||||
|
CD40L_MOUSE
|
||||||
| θ value | 6.88961 (rank : 56) | NC score | 0.025098 (rank : 58) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27548 | Gene names | Cd40lg, Cd40l, Tnfsf5 | |||
|
Domain Architecture |
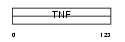
|
|||||
| Description | CD40 ligand (CD40-L) (Tumor necrosis factor ligand superfamily member 5) (CD154 antigen) (TNF-related activation protein) (TRAP) (T cell antigen Gp39) [Contains: CD40 ligand, membrane form; CD40 ligand, soluble form]. | |||||
|
CP135_MOUSE
|
||||||
| θ value | 6.88961 (rank : 57) | NC score | 0.026722 (rank : 52) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
DESP_HUMAN
|
||||||
| θ value | 6.88961 (rank : 58) | NC score | 0.024164 (rank : 60) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 59) | NC score | 0.028452 (rank : 47) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
MY18A_MOUSE
|
||||||
| θ value | 6.88961 (rank : 60) | NC score | 0.031319 (rank : 38) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 6.88961 (rank : 61) | NC score | 0.023737 (rank : 61) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 6.88961 (rank : 62) | NC score | 0.028529 (rank : 46) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
REST_MOUSE
|
||||||
| θ value | 6.88961 (rank : 63) | NC score | 0.026147 (rank : 54) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
STABP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 64) | NC score | 0.021014 (rank : 64) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CQ26, Q3UTI9 | Gene names | Stambp, Amsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STAM-binding protein (EC 3.1.2.15) (Associated molecule with the SH3 domain of STAM). | |||||
|
SYCP1_MOUSE
|
||||||
| θ value | 6.88961 (rank : 65) | NC score | 0.028425 (rank : 48) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
TAF3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 66) | NC score | 0.024297 (rank : 59) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
TCEA3_HUMAN
|
||||||
| θ value | 6.88961 (rank : 67) | NC score | 0.016915 (rank : 66) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75764 | Gene names | TCEA3, TFIISH | |||
|
Domain Architecture |
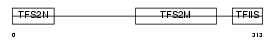
|
|||||
| Description | Transcription elongation factor A protein 3 (Transcription elongation factor S-II protein 3) (Transcription elongation factor TFIIS.h). | |||||
|
ZN740_HUMAN
|
||||||
| θ value | 6.88961 (rank : 68) | NC score | 0.006587 (rank : 75) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 684 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NDX6 | Gene names | ZNF740, TB7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 740 (OriLyt TD-element-binding protein 7). | |||||
|
ZN740_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.006601 (rank : 74) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6NZQ6, Q3UMS3, Q8JZR3 | Gene names | Znf740, Zfp740 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 740. | |||||
|
CLC10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 70) | NC score | 0.007177 (rank : 73) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IUN9, Q14538, Q6PIW3 | Gene names | CLEC10A, CLECSF14, HML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 10 member A (C-type lectin superfamily member 14) (Macrophage lectin 2) (CD301 antigen). | |||||
|
ERBB3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 71) | NC score | 0.000721 (rank : 80) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P21860 | Gene names | ERBB3, HER3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Tyrosine kinase-type cell surface receptor HER3). | |||||
|
FGD4_MOUSE
|
||||||
| θ value | 8.99809 (rank : 72) | NC score | 0.007463 (rank : 72) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
ITAL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 73) | NC score | 0.006432 (rank : 76) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20701, O43746, Q45H73, Q9UBC8 | Gene names | ITGAL, CD11A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-L precursor (Leukocyte adhesion glycoprotein LFA-1 alpha chain) (LFA-1A) (Leukocyte function-associated molecule 1 alpha chain) (CD11a antigen). | |||||
|
K1H2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 74) | NC score | 0.016457 (rank : 67) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62168 | Gene names | Krtha2, Hka2, Krt1-2 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
MAP9_MOUSE
|
||||||
| θ value | 8.99809 (rank : 75) | NC score | 0.032232 (rank : 35) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
RSF1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 76) | NC score | 0.030153 (rank : 42) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
SARM1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 77) | NC score | 0.051961 (rank : 24) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6SZW1, O60277, Q7LGG3, Q9NXY5 | Gene names | SARM1, KIAA0524, SARM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Sterile alpha and Armadillo repeat protein) (Tir-1 homolog). | |||||
|
SC11A_MOUSE
|
||||||
| θ value | 8.99809 (rank : 78) | NC score | 0.005652 (rank : 77) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
SCG1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.021356 (rank : 63) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16014 | Gene names | Chgb, Scg-1, Scg1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB). | |||||
|
SCML1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.051311 (rank : 25) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UN30 | Gene names | SCML1 | |||
|
Domain Architecture |

|
|||||
| Description | Sex comb on midleg-like protein 1. | |||||
|
SAMD9_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 79 | |
| SwissProt Accessions | Q5K651, Q6P080, Q75N21, Q8IVG6, Q9NXS8 | Gene names | SAMD9, KIAA2004, OEF1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 9. | |||||
|
SHAN2_HUMAN
|
||||||
| NC score | 0.221531 (rank : 2) | θ value | 6.43352e-06 (rank : 2) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 346 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9UPX8, Q3Y8G9, Q52LK2, Q9UKP1 | Gene names | SHANK2, KIAA1022 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2). | |||||
|
SHAN2_MOUSE
|
||||||
| NC score | 0.210563 (rank : 3) | θ value | 6.43352e-06 (rank : 3) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q80Z38, Q3UTK4, Q5DU07 | Gene names | Shank2, Kiaa1022 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 2 (Shank2) (Cortactin- binding protein 1) (CortBP1). | |||||
|
PHC1_HUMAN
|
||||||
| NC score | 0.172095 (rank : 4) | θ value | 0.00869519 (rank : 5) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P78364, Q8WVM3, Q9BU63 | Gene names | PHC1, EDR1, PH1 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (hPH1) (Early development regulatory protein 1). | |||||
|
PHC1_MOUSE
|
||||||
| NC score | 0.169301 (rank : 5) | θ value | 0.00509761 (rank : 4) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q64028, P70359, Q64307, Q8BZ80 | Gene names | Phc1, Edr, Edr1, Rae28 | |||
|
Domain Architecture |

|
|||||
| Description | Polyhomeotic-like protein 1 (mPH1) (Early development regulatory protein 1) (RAE-28). | |||||
|
PHC2_MOUSE
|
||||||
| NC score | 0.163440 (rank : 6) | θ value | 0.0193708 (rank : 7) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9QWH1, O88463, Q8K5D9 | Gene names | Phc2, Edr2, Ph2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (mPH2) (Early development regulatory protein 2) (p36). | |||||
|
PHC2_HUMAN
|
||||||
| NC score | 0.153208 (rank : 7) | θ value | 0.0193708 (rank : 6) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 388 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IXK0, Q5T0C1, Q6NUJ6, Q6ZQR1, Q8N306, Q8TAG8, Q96BL4, Q9Y4Y7 | Gene names | PHC2, EDR2, PH2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 2 (hPH2) (Early development regulatory protein 2). | |||||
|
PHC3_HUMAN
|
||||||
| NC score | 0.129003 (rank : 8) | θ value | 0.47712 (rank : 17) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NDX5, Q5HYF0, Q6NSG2, Q8NFT7, Q8NFZ1, Q8TBM2, Q9H971, Q9H9I4 | Gene names | PHC3, EDR3, PH3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3 (hPH3) (Homolog of polyhomeotic 3) (Early development regulatory protein 3). | |||||
|
PHC3_MOUSE
|
||||||
| NC score | 0.123773 (rank : 9) | θ value | 1.38821 (rank : 28) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8CHP6, Q3TLV5 | Gene names | Phc3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyhomeotic-like protein 3. | |||||
|
SAM14_HUMAN
|
||||||
| NC score | 0.118797 (rank : 10) | θ value | 1.06291 (rank : 25) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 81 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8IZD0, Q8N2X0 | Gene names | SAMD14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 14. | |||||
|
SAM14_MOUSE
|
||||||
| NC score | 0.114134 (rank : 11) | θ value | 0.813845 (rank : 22) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 91 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q8K070, Q5SWB7, Q8BHE2, Q8C8N5 | Gene names | Samd14 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha motif domain-containing protein 14. | |||||
|
NOP56_MOUSE
|
||||||
| NC score | 0.089265 (rank : 12) | θ value | 0.163984 (rank : 9) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 129 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9D6Z1, Q3UD45, Q8BVL1, Q8VDT2, Q99LT8, Q9CT15 | Gene names | Nol5a, Nop56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
MGAT5_MOUSE
|
||||||
| NC score | 0.084596 (rank : 13) | θ value | 0.47712 (rank : 15) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8R4G6 | Gene names | Mgat5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V (EC 2.4.1.155) (Mannoside acetylglucosaminyltransferase 5) (Alpha- mannoside beta-1,6-N-acetylglucosaminyltransferase) (N- acetylglucosaminyl-transferase V) (GNT-V) (GlcNAc-T V). | |||||
|
MGAT5_HUMAN
|
||||||
| NC score | 0.082682 (rank : 14) | θ value | 0.62314 (rank : 18) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q09328 | Gene names | MGAT5, GGNT5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V (EC 2.4.1.155) (Mannoside acetylglucosaminyltransferase 5) (Alpha- mannoside beta-1,6-N-acetylglucosaminyltransferase) (N- acetylglucosaminyl-transferase V) (GNT-V) (GlcNAc-T V). | |||||
|
NOP56_HUMAN
|
||||||
| NC score | 0.081208 (rank : 15) | θ value | 0.47712 (rank : 16) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 95 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O00567, Q9NQ05 | Gene names | NOL5A, NOP56 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar protein Nop56 (Nucleolar protein 5A). | |||||
|
SHAN1_HUMAN
|
||||||
| NC score | 0.080451 (rank : 16) | θ value | 0.0252991 (rank : 8) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1041 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9Y566, Q9NYW9 | Gene names | SHANK1 | |||
|
Domain Architecture |

|
|||||
| Description | SH3 and multiple ankyrin repeat domains protein 1 (Shank1) (Somatostatin receptor-interacting protein) (SSTR-interacting protein) (SSTRIP). | |||||
|
IFIT2_HUMAN
|
||||||
| NC score | 0.069732 (rank : 17) | θ value | 0.365318 (rank : 13) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 53 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P09913 | Gene names | IFIT2, G10P2, IFI54 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 2 (IFIT-2) (Interferon-induced 54 kDa protein) (IFI-54K) (ISG-54 K). | |||||
|
IFIT2_MOUSE
|
||||||
| NC score | 0.069472 (rank : 18) | θ value | 0.47712 (rank : 14) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q64112, Q62385 | Gene names | Ifit2, Garg39, Ifi54 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 2 (IFIT-2) (Interferon-induced 54 kDa protein) (IFI-54K) (Glucocorticoid- attenuated response gene 39 protein) (GARG-39). | |||||
|
CF066_MOUSE
|
||||||
| NC score | 0.067567 (rank : 19) | θ value | 1.06291 (rank : 24) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9D1H6, Q3TV72 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | UPF0240 protein C6orf66 homolog. | |||||
|
NEB1_HUMAN
|
||||||
| NC score | 0.065129 (rank : 20) | θ value | 0.21417 (rank : 12) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 829 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | Q9ULJ8, O76059, Q9NXT2 | Gene names | PPP1R9A, KIAA1222 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neurabin-1 (Neurabin-I) (Neural tissue-specific F-actin-binding protein I) (Protein phosphatase 1 regulatory subunit 9A). | |||||
|
MPP10_MOUSE
|
||||||
| NC score | 0.054389 (rank : 21) | θ value | 0.813845 (rank : 20) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q810V0 | Gene names | Mphosph10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | U3 small nucleolar ribonucleoprotein protein MPP10 (M phase phosphoprotein 10). | |||||
|
IFIT3_HUMAN
|
||||||
| NC score | 0.054155 (rank : 22) | θ value | 2.36792 (rank : 36) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 83 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14879, Q99634, Q9BSK7 | Gene names | IFIT3, IFI60, IFIT4 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced protein with tetratricopeptide repeats 3 (IFIT-3) (IFIT-4) (Interferon-induced 60 kDa protein) (IFI-60K) (ISG-60) (CIG49) (Retinoic acid-induced gene G protein) (RIG-G). | |||||
|
MAP1B_MOUSE
|
||||||
| NC score | 0.054113 (rank : 23) | θ value | 0.21417 (rank : 11) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 977 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | P14873 | Gene names | Map1b, Mtap1b, Mtap5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 1B (MAP 1B) (MAP1.2) (MAP1(X)) [Contains: MAP1 light chain LC1]. | |||||
|
SARM1_HUMAN
|
||||||
| NC score | 0.051961 (rank : 24) | θ value | 8.99809 (rank : 77) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 58 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q6SZW1, O60277, Q7LGG3, Q9NXY5 | Gene names | SARM1, KIAA0524, SARM | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sterile alpha and TIR motif-containing protein 1 (Sterile alpha and Armadillo repeat protein) (Tir-1 homolog). | |||||
|
SCML1_HUMAN
|
||||||
| NC score | 0.051311 (rank : 25) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UN30 | Gene names | SCML1 | |||
|
Domain Architecture |

|
|||||
| Description | Sex comb on midleg-like protein 1. | |||||
|
DEK_MOUSE
|
||||||
| NC score | 0.046961 (rank : 26) | θ value | 2.36792 (rank : 34) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 280 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q7TNV0, Q80VC5, Q8BZV6 | Gene names | Dek | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein DEK. | |||||
|
RBBP6_HUMAN
|
||||||
| NC score | 0.042845 (rank : 27) | θ value | 1.38821 (rank : 29) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1135 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q7Z6E9, Q15290, Q6DKH4, Q6P4C2, Q6YNC9, Q7Z6E8, Q8N0V2, Q96PH3, Q9H3I8, Q9H5M5, Q9NPX4 | Gene names | RBBP6, P2PR, PACT, RBQ1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Retinoblastoma-binding protein 6 (p53-associated cellular protein of testis) (Proliferation potential-related protein) (Protein P2P-R) (Retinoblastoma-binding Q protein 1) (Protein RBQ-1). | |||||
|
SFRIP_HUMAN
|
||||||
| NC score | 0.042013 (rank : 28) | θ value | 3.0926 (rank : 42) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 530 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q99590, Q8IW59 | Gene names | SFRS2IP, CASP11, SIP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | SFRS2-interacting protein (Splicing factor, arginine/serine-rich 2- interacting protein) (SC35-interacting protein 1) (CTD-associated SR protein 11) (Splicing regulatory protein 129) (SRrp129) (NY-REN-40 antigen). | |||||
|
AREG_MOUSE
|
||||||
| NC score | 0.040051 (rank : 29) | θ value | 3.0926 (rank : 37) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 40 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P31955 | Gene names | Areg, Sdgf | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Amphiregulin precursor (AR) (Schwannoma-derived growth factor) (SDGF). | |||||
|
ZN291_HUMAN
|
||||||
| NC score | 0.039069 (rank : 30) | θ value | 3.0926 (rank : 45) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 647 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q9BY12 | Gene names | ZNF291 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 291. | |||||
|
GOGA1_MOUSE
|
||||||
| NC score | 0.038389 (rank : 31) | θ value | 0.21417 (rank : 10) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 994 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q9CW79, Q80YB0 | Gene names | Golga1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 1 (Golgin-97). | |||||
|
CE290_HUMAN
|
||||||
| NC score | 0.035642 (rank : 32) | θ value | 1.06291 (rank : 23) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1553 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | O15078, Q1PSK5, Q66GS8, Q9H2G6, Q9H6Q7, Q9H8I0 | Gene names | CEP290, KIAA0373, NPHP6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein Cep290 (Nephrocystin-6) (Tumor antigen se2-2). | |||||
|
AFF4_MOUSE
|
||||||
| NC score | 0.033765 (rank : 33) | θ value | 1.81305 (rank : 30) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 581 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q9ESC8, Q8C6K4, Q8C6W3, Q8CCH3 | Gene names | Aff4, Alf4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | AF4/FMR2 family member 4. | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.032441 (rank : 34) | θ value | 1.38821 (rank : 26) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MAP9_MOUSE
|
||||||
| NC score | 0.032232 (rank : 35) | θ value | 8.99809 (rank : 75) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 529 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | Q3TRR0, Q3UUD1, Q3UX85, Q3UXE7, Q5M8N8, Q6P8K1, Q8BMM4, Q8BYP7 | Gene names | Map9, Asap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Microtubule-associated protein 9 (Aster-associated protein). | |||||
|
MYO5A_HUMAN
|
||||||
| NC score | 0.031652 (rank : 36) | θ value | 0.62314 (rank : 19) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1092 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | Q9Y4I1, O60653, Q07902, Q16249, Q9UE30, Q9UE31 | Gene names | MYO5A, MYH12 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle) (Myosin heavy chain 12) (Myoxin). | |||||
|
RBCC1_MOUSE
|
||||||
| NC score | 0.031319 (rank : 37) | θ value | 3.0926 (rank : 41) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1048 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q9ESK9, Q3TXX2, Q61384, Q8BRY9, Q9JK14 | Gene names | Rb1cc1, Cc1, Kiaa0203 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | RB1-inducible coiled-coil protein 1 (Coiled-coil-forming protein 1) (LaXp180 protein). | |||||
|
MY18A_MOUSE
|
||||||
| NC score | 0.031319 (rank : 38) | θ value | 6.88961 (rank : 60) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1989 | Shared Neighborhood Hits | 42 | |
| SwissProt Accessions | Q9JMH9, Q811D7 | Gene names | Myo18a, Myspdz | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-18A (Myosin XVIIIa) (Myosin containing PDZ domain) (Molecule associated with JAK3 N-terminus) (MAJN). | |||||
|
TCOF_HUMAN
|
||||||
| NC score | 0.031243 (rank : 39) | θ value | 4.03905 (rank : 47) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 543 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q13428, Q99408, Q99860 | Gene names | TCOF1 | |||
|
Domain Architecture |

|
|||||
| Description | Treacle protein (Treacher Collins syndrome protein). | |||||
|
ZRF1_HUMAN
|
||||||
| NC score | 0.030841 (rank : 40) | θ value | 3.0926 (rank : 46) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 457 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q99543 | Gene names | ZRF1, DNAJC2, MPHOSPH11, MPP11 | |||
|
Domain Architecture |

|
|||||
| Description | Zuotin-related factor 1 (M-phase phosphoprotein 11). | |||||
|
GBP2_HUMAN
|
||||||
| NC score | 0.030780 (rank : 41) | θ value | 2.36792 (rank : 35) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 349 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | P32456, Q86TB0 | Gene names | GBP2 | |||
|
Domain Architecture |

|
|||||
| Description | Interferon-induced guanylate-binding protein 2 (GTP-binding protein 2) (Guanine nucleotide-binding protein 2) (GBP-2) (HuGBP-2). | |||||
|
RSF1_HUMAN
|
||||||
| NC score | 0.030153 (rank : 42) | θ value | 8.99809 (rank : 76) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 539 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q96T23, Q86X86, Q9H3L8, Q9NVZ8, Q9NYU0 | Gene names | RSF1, HBXAP, XAP8 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Remodeling and spacing factor 1 (Rsf-1) (Hepatitis B virus X- associated protein) (HBV pX-associated protein 8) (p325 subunit of RSF chromatin remodelling complex). | |||||
|
MYO5A_MOUSE
|
||||||
| NC score | 0.029802 (rank : 43) | θ value | 1.38821 (rank : 27) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1117 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q99104 | Gene names | Myo5a, Dilute | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-5A (Myosin Va) (Dilute myosin heavy chain, non-muscle). | |||||
|
GOGB1_HUMAN
|
||||||
| NC score | 0.028960 (rank : 44) | θ value | 5.27518 (rank : 52) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 2297 | Shared Neighborhood Hits | 41 | |
| SwissProt Accessions | Q14789, Q14398 | Gene names | GOLGB1 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily B member 1 (Giantin) (Macrogolgin) (372 kDa Golgi complex-associated protein) (GCP372). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.028708 (rank : 45) | θ value | 1.81305 (rank : 32) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 29 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.028529 (rank : 46) | θ value | 6.88961 (rank : 62) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.028452 (rank : 47) | θ value | 6.88961 (rank : 59) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
SYCP1_MOUSE
|
||||||
| NC score | 0.028425 (rank : 48) | θ value | 6.88961 (rank : 65) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1456 | Shared Neighborhood Hits | 33 | |
| SwissProt Accessions | Q62209, O09205, P70192, Q62329 | Gene names | Sycp1, Scp1 | |||
|
Domain Architecture |

|
|||||
| Description | Synaptonemal complex protein 1 (SCP-1). | |||||
|
NFM_MOUSE
|
||||||
| NC score | 0.028398 (rank : 49) | θ value | 0.813845 (rank : 21) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1159 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | P08553, Q61961 | Gene names | Nef3, Nefm, Nfm | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M). | |||||
|
CS007_MOUSE
|
||||||
| NC score | 0.027794 (rank : 50) | θ value | 5.27518 (rank : 51) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 464 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q6ZPZ3 | Gene names | Kiaa1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7 homolog. | |||||
|
AP3B1_MOUSE
|
||||||
| NC score | 0.027605 (rank : 51) | θ value | 1.81305 (rank : 31) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 302 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9Z1T1, Q91YR4 | Gene names | Ap3b1 | |||
|
Domain Architecture |

|
|||||
| Description | AP-3 complex subunit beta-1 (Adapter-related protein complex 3 beta-1 subunit) (Beta3A-adaptin) (Adaptor protein complex AP-3 beta-1 subunit) (Clathrin assembly protein complex 3 beta-1 large chain). | |||||
|
CP135_MOUSE
|
||||||
| NC score | 0.026722 (rank : 52) | θ value | 6.88961 (rank : 57) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1278 | Shared Neighborhood Hits | 28 | |
| SwissProt Accessions | Q6P5D4, Q6A030 | Gene names | Cep135, Cep4, Kiaa0635 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosomal protein of 135 kDa (Cep135 protein) (Centrosomal protein 4). | |||||
|
NFM_HUMAN
|
||||||
| NC score | 0.026247 (rank : 53) | θ value | 3.0926 (rank : 40) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1328 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | P07197 | Gene names | NEF3, NEFM, NFM | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet M protein (160 kDa neurofilament protein) (Neurofilament medium polypeptide) (NF-M) (Neurofilament 3). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.026147 (rank : 54) | θ value | 6.88961 (rank : 63) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
CS007_HUMAN
|
||||||
| NC score | 0.026056 (rank : 55) | θ value | 5.27518 (rank : 50) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 401 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UPT8, Q9Y420 | Gene names | C19orf7, KIAA1064 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger CCCH domain-containing protein C19orf7. | |||||
|
CC019_HUMAN
|
||||||
| NC score | 0.025734 (rank : 56) | θ value | 6.88961 (rank : 55) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6PII3, Q96CS5 | Gene names | C3orf19 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C3orf19. | |||||
|
MFN1_MOUSE
|
||||||
| NC score | 0.025487 (rank : 57) | θ value | 3.0926 (rank : 39) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 55 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q811U4, Q811D5, Q8CEY6, Q99M10, Q9D395 | Gene names | Mfn1 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane GTPase MFN1 (EC 3.6.5.-) (Mitofusin-1). | |||||
|
CD40L_MOUSE
|
||||||
| NC score | 0.025098 (rank : 58) | θ value | 6.88961 (rank : 56) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P27548 | Gene names | Cd40lg, Cd40l, Tnfsf5 | |||
|
Domain Architecture |

|
|||||
| Description | CD40 ligand (CD40-L) (Tumor necrosis factor ligand superfamily member 5) (CD154 antigen) (TNF-related activation protein) (TRAP) (T cell antigen Gp39) [Contains: CD40 ligand, membrane form; CD40 ligand, soluble form]. | |||||
|
TAF3_HUMAN
|
||||||
| NC score | 0.024297 (rank : 59) | θ value | 6.88961 (rank : 66) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 568 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q5VWG9, Q6GMS5, Q6P6B5, Q86VY6, Q9BQS9, Q9UFI8 | Gene names | TAF3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transcription initiation factor TFIID subunit 3 (TBP-associated factor 3) (Transcription initiation factor TFIID 140 kDa subunit) (140 kDa TATA box-binding protein-associated factor) (TAF140) (TAFII140). | |||||
|
DESP_HUMAN
|
||||||
| NC score | 0.024164 (rank : 60) | θ value | 6.88961 (rank : 58) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1470 | Shared Neighborhood Hits | 30 | |
| SwissProt Accessions | P15924, O75993, Q14189, Q9UHN4 | Gene names | DSP | |||
|
Domain Architecture |

|
|||||
| Description | Desmoplakin (DP) (250/210 kDa paraneoplastic pemphigus antigen). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.023737 (rank : 61) | θ value | 6.88961 (rank : 61) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
TIAM1_MOUSE
|
||||||
| NC score | 0.022283 (rank : 62) | θ value | 4.03905 (rank : 48) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 205 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q60610 | Gene names | Tiam1, Tiam-1 | |||
|
Domain Architecture |

|
|||||
| Description | T-lymphoma invasion and metastasis-inducing protein 1 (TIAM-1 protein). | |||||
|
SCG1_MOUSE
|
||||||
| NC score | 0.021356 (rank : 63) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | P16014 | Gene names | Chgb, Scg-1, Scg1 | |||
|
Domain Architecture |

|
|||||
| Description | Secretogranin-1 precursor (Secretogranin I) (SgI) (Chromogranin B) (CgB). | |||||
|
STABP_MOUSE
|
||||||
| NC score | 0.021014 (rank : 64) | θ value | 6.88961 (rank : 64) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q9CQ26, Q3UTI9 | Gene names | Stambp, Amsh | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | STAM-binding protein (EC 3.1.2.15) (Associated molecule with the SH3 domain of STAM). | |||||
|
BRDT_MOUSE
|
||||||
| NC score | 0.019928 (rank : 65) | θ value | 3.0926 (rank : 38) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q91Y44, Q59HJ4 | Gene names | Brdt, Fsrg3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Bromodomain testis-specific protein (RING3-like protein) (Bromodomain- containing female sterile homeotic-like protein). | |||||
|
TCEA3_HUMAN
|
||||||
| NC score | 0.016915 (rank : 66) | θ value | 6.88961 (rank : 67) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75764 | Gene names | TCEA3, TFIISH | |||
|
Domain Architecture |

|
|||||
| Description | Transcription elongation factor A protein 3 (Transcription elongation factor S-II protein 3) (Transcription elongation factor TFIIS.h). | |||||
|
K1H2_MOUSE
|
||||||
| NC score | 0.016457 (rank : 67) | θ value | 8.99809 (rank : 74) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 446 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q62168 | Gene names | Krtha2, Hka2, Krt1-2 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type I cuticular Ha2 (Hair keratin, type I Ha2). | |||||
|
ANKR5_MOUSE
|
||||||
| NC score | 0.016332 (rank : 68) | θ value | 2.36792 (rank : 33) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 298 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D2J7 | Gene names | Ankrd5 | |||
|
Domain Architecture |

|
|||||
| Description | Ankyrin repeat domain-containing protein 5. | |||||
|
TSH2_MOUSE
|
||||||
| NC score | 0.014680 (rank : 69) | θ value | 5.27518 (rank : 53) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 467 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q68FE9, Q5DTF1, Q6AXH5, Q9JL71 | Gene names | Tshz2, Kiaa4248, Sdccag33l, Tsh2, Zfp218, Znf218 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 2 (Zinc finger protein 218) (SDCCAG33-like protein). | |||||
|
ZN148_MOUSE
|
||||||
| NC score | 0.008708 (rank : 70) | θ value | 3.0926 (rank : 44) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 755 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q61624, P97475 | Gene names | Znf148, Zbp89, Zfp148 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89) (G-rich box-binding protein) (Beta enolase repressor factor 1) (Transcription factor BFCOL1). | |||||
|
ZN148_HUMAN
|
||||||
| NC score | 0.008619 (rank : 71) | θ value | 3.0926 (rank : 43) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 747 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9UQR1, O00389, O43591 | Gene names | ZNF148, ZBP89 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 148 (Zinc finger DNA-binding protein 89) (Transcription factor ZBP-89). | |||||
|
FGD4_MOUSE
|
||||||
| NC score | 0.007463 (rank : 72) | θ value | 8.99809 (rank : 72) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 227 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91ZT5, Q3UEB6, Q8BW60, Q8BZI7, Q91ZT3, Q91ZT4 | Gene names | Fgd4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE, RhoGEF and PH domain-containing protein 4 (Actin filament- binding protein frabin) (FGD1-related F-actin-binding protein). | |||||
|
CLC10_HUMAN
|
||||||
| NC score | 0.007177 (rank : 73) | θ value | 8.99809 (rank : 70) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 134 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8IUN9, Q14538, Q6PIW3 | Gene names | CLEC10A, CLECSF14, HML | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | C-type lectin domain family 10 member A (C-type lectin superfamily member 14) (Macrophage lectin 2) (CD301 antigen). | |||||
|
ZN740_MOUSE
|
||||||
| NC score | 0.006601 (rank : 74) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 685 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q6NZQ6, Q3UMS3, Q8JZR3 | Gene names | Znf740, Zfp740 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 740. | |||||
|
ZN740_HUMAN
|
||||||
| NC score | 0.006587 (rank : 75) | θ value | 6.88961 (rank : 68) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 684 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8NDX6 | Gene names | ZNF740, TB7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 740 (OriLyt TD-element-binding protein 7). | |||||
|
ITAL_HUMAN
|
||||||
| NC score | 0.006432 (rank : 76) | θ value | 8.99809 (rank : 73) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P20701, O43746, Q45H73, Q9UBC8 | Gene names | ITGAL, CD11A | |||
|
Domain Architecture |

|
|||||
| Description | Integrin alpha-L precursor (Leukocyte adhesion glycoprotein LFA-1 alpha chain) (LFA-1A) (Leukocyte function-associated molecule 1 alpha chain) (CD11a antigen). | |||||
|
SC11A_MOUSE
|
||||||
| NC score | 0.005652 (rank : 77) | θ value | 8.99809 (rank : 78) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9R053, Q9JMD4 | Gene names | Scn11a, Nan, Nat, Sns2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Sodium channel protein type 11 subunit alpha (Sodium channel protein type XI subunit alpha) (Voltage-gated sodium channel subunit alpha Nav1.9) (Sensory neuron sodium channel 2) (NaN). | |||||
|
ZN484_HUMAN
|
||||||
| NC score | 0.002532 (rank : 78) | θ value | 4.03905 (rank : 49) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 766 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q5JVG2 | Gene names | ZNF484 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 484. | |||||
|
ZN485_HUMAN
|
||||||
| NC score | 0.002362 (rank : 79) | θ value | 5.27518 (rank : 54) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 744 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8NCK3, Q96CL0 | Gene names | ZNF485 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Zinc finger protein 485. | |||||
|
ERBB3_HUMAN
|
||||||
| NC score | 0.000721 (rank : 80) | θ value | 8.99809 (rank : 71) | |||
| Query Neighborhood Hits | 79 | Target Neighborhood Hits | 918 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P21860 | Gene names | ERBB3, HER3 | |||
|
Domain Architecture |

|
|||||
| Description | Receptor tyrosine-protein kinase erbB-3 precursor (EC 2.7.10.1) (c- erbB3) (Tyrosine kinase-type cell surface receptor HER3). | |||||