Please be patient as the page loads
|
PRGB_HUMAN
|
||||||
| SwissProt Accessions | Q02325 | Gene names | PLGL, PRGB | |||
|
Domain Architecture |
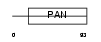
|
|||||
| Description | Plasminogen-related protein B precursor. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PRGB_HUMAN
|
||||||
| θ value | 5.96599e-36 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02325 | Gene names | PLGL, PRGB | |||
|
Domain Architecture |

|
|||||
| Description | Plasminogen-related protein B precursor. | |||||
|
PLMN_HUMAN
|
||||||
| θ value | 1.24399e-33 (rank : 2) | NC score | 0.246510 (rank : 2) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P00747, Q15146, Q6PA00 | Gene names | PLG | |||
|
Domain Architecture |

|
|||||
| Description | Plasminogen precursor (EC 3.4.21.7) [Contains: Plasmin heavy chain A; Activation peptide; Angiostatin; Plasmin heavy chain A, short form; Plasmin light chain B]. | |||||
|
PLMN_MOUSE
|
||||||
| θ value | 7.54701e-31 (rank : 3) | NC score | 0.244021 (rank : 3) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20918, Q8CIS2, Q91WJ5 | Gene names | Plg | |||
|
Domain Architecture |

|
|||||
| Description | Plasminogen precursor (EC 3.4.21.7) [Contains: Plasmin heavy chain A; Activation peptide; Angiostatin; Plasmin heavy chain A, short form; Plasmin light chain B]. | |||||
|
HGF_HUMAN
|
||||||
| θ value | 0.62314 (rank : 4) | NC score | 0.199178 (rank : 4) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P14210, Q02935, Q13494, Q14519, Q8TCE2, Q9BYL9, Q9BYM0, Q9UDU6 | Gene names | HGF, HPTA | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor precursor (Scatter factor) (SF) (Hepatopoeitin A) [Contains: Hepatocyte growth factor alpha chain; Hepatocyte growth factor beta chain]. | |||||
|
BAP29_HUMAN
|
||||||
| θ value | 3.0926 (rank : 5) | NC score | 0.038022 (rank : 180) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UHQ4, O95003 | Gene names | BCAP29, BAP29 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor-associated protein 29 (BCR-associated protein Bap29). | |||||
|
HGF_MOUSE
|
||||||
| θ value | 4.03905 (rank : 6) | NC score | 0.196779 (rank : 5) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08048, Q61662, Q64007, Q6LBE6 | Gene names | Hgf | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor precursor (Scatter factor) (SF) (Hepatopoeitin A) [Contains: Hepatocyte growth factor alpha chain; Hepatocyte growth factor beta chain]. | |||||
|
ACRO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 7) | NC score | 0.102725 (rank : 54) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10323, Q6ICK2 | Gene names | ACR, ACRS | |||
|
Domain Architecture |
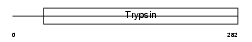
|
|||||
| Description | Acrosin precursor (EC 3.4.21.10) [Contains: Acrosin light chain; Acrosin heavy chain]. | |||||
|
ACRO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 8) | NC score | 0.104653 (rank : 42) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23578 | Gene names | Acr | |||
|
Domain Architecture |
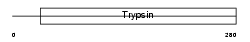
|
|||||
| Description | Acrosin precursor (EC 3.4.21.10) [Contains: Acrosin light chain; Acrosin heavy chain]. | |||||
|
APOA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 9) | NC score | 0.190519 (rank : 6) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08519 | Gene names | LPA | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein(a) precursor (EC 3.4.21.-) (Apo(a)) (Lp(a)). | |||||
|
BSSP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 10) | NC score | 0.101466 (rank : 61) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZN4, O43342, Q6UXE0 | Gene names | PRSS22, BSSP4, PRSS26 | |||
|
Domain Architecture |
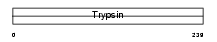
|
|||||
| Description | Brain-specific serine protease 4 precursor (EC 3.4.21.-) (BSSP-4) (Serine protease 22) (Tryptase epsilon). | |||||
|
BSSP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 11) | NC score | 0.099605 (rank : 70) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ER10 | Gene names | Prss22, Bssp4, Prss26 | |||
|
Domain Architecture |
|
|||||
| Description | Brain-specific serine protease 4 precursor (EC 3.4.21.-) (BSSP-4) (Serine protease 22) (Tryptase epsilon). | |||||
|
C1RA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 12) | NC score | 0.079045 (rank : 171) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG16, Q99KI6, Q9ET60 | Gene names | C1ra, C1r | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r-A subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-A subcomponent) [Contains: Complement C1r-A subcomponent heavy chain; Complement C1r-A subcomponent light chain]. | |||||
|
C1RB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 13) | NC score | 0.078113 (rank : 172) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CFG9 | Gene names | C1rb, C1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1r-B subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-B subcomponent) [Contains: Complement C1r-B subcomponent heavy chain; Complement C1r-B subcomponent light chain]. | |||||
|
C1R_HUMAN
|
||||||
| θ value | θ > 10 (rank : 14) | NC score | 0.077524 (rank : 173) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00736, Q8J012 | Gene names | C1R | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r subcomponent precursor (EC 3.4.21.41) (Complement component 1, r subcomponent) [Contains: Complement C1r subcomponent heavy chain; Complement C1r subcomponent light chain]. | |||||
|
C1S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 15) | NC score | 0.085253 (rank : 158) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09871, Q9UCU7, Q9UCU8, Q9UCU9, Q9UCV0, Q9UCV1, Q9UCV2, Q9UCV3, Q9UCV4, Q9UCV5, Q9UM14 | Gene names | C1S | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1s subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s subcomponent heavy chain; Complement C1s subcomponent light chain]. | |||||
|
CAP7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 16) | NC score | 0.101030 (rank : 64) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20160, P80014, Q52LG4 | Gene names | AZU1 | |||
|
Domain Architecture |
|
|||||
| Description | Azurocidin precursor (Cationic antimicrobial protein CAP37) (Heparin- binding protein) (HBP). | |||||
|
CATG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 17) | NC score | 0.091313 (rank : 117) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08311 | Gene names | CTSG | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin G precursor (EC 3.4.21.20) (CG). | |||||
|
CATG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 18) | NC score | 0.096566 (rank : 90) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28293 | Gene names | Ctsg | |||
|
Domain Architecture |
|
|||||
| Description | Cathepsin G precursor (EC 3.4.21.20) (Vimentin-specific protease) (VSP). | |||||
|
CFAB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 19) | NC score | 0.071290 (rank : 174) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00751, O15006, Q29944, Q96HX6, Q9BTF5, Q9BX92 | Gene names | CFB, BF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) (Properdin factor B) (Glycine-rich beta glycoprotein) (GBG) (PBF2) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
CFAB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 20) | NC score | 0.070602 (rank : 175) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04186 | Gene names | Cfb, Bf, H2-Bf | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
CFAD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 21) | NC score | 0.094298 (rank : 102) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00746 | Gene names | CFD, DF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor D precursor (EC 3.4.21.46) (C3 convertase activator) (Properdin factor D) (Adipsin). | |||||
|
CFAD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 22) | NC score | 0.094907 (rank : 97) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P03953, Q61280 | Gene names | Cfd, Adn, Df | |||
|
Domain Architecture |
|
|||||
| Description | Complement factor D precursor (EC 3.4.21.46) (C3 convertase activator) (Properdin factor D) (Adipsin) (28 kDa adipocyte protein). | |||||
|
CFAI_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.092152 (rank : 111) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05156, O60442 | Gene names | CFI, IF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
CFAI_MOUSE
|
||||||
| θ value | θ > 10 (rank : 24) | NC score | 0.089131 (rank : 137) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
CLCR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.105086 (rank : 37) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99895, O00765, Q9NUH5 | Gene names | CTRC, CLCR | |||
|
Domain Architecture |
|
|||||
| Description | Caldecrin precursor (EC 3.4.21.2) (Chymotrypsin C). | |||||
|
CO2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.056916 (rank : 177) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06681, O19694, Q13904 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
CO2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.060499 (rank : 176) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21180 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
CORIN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.084970 (rank : 161) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5Q5, Q9UHY2 | Gene names | CORIN, CRN, TMPRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Heart-specific serine proteinase ATC2) (Transmembrane protease, serine 10). | |||||
|
CORIN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.085051 (rank : 160) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
CS1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.086709 (rank : 148) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG14, Q8BJC4, Q8CH28, Q8VBY4 | Gene names | C1sa, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-A subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-A subcomponent heavy chain; Complement C1s-A subcomponent light chain]. | |||||
|
CS1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.085501 (rank : 156) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CFG8 | Gene names | C1sb, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-B subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-B subcomponent heavy chain; Complement C1s-B subcomponent light chain]. | |||||
|
CTRB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.104254 (rank : 48) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17538 | Gene names | CTRB1, CTRB | |||
|
Domain Architecture |
|
|||||
| Description | Chymotrypsinogen B precursor (EC 3.4.21.1) [Contains: Chymotrypsin B chain A; Chymotrypsin B chain B; Chymotrypsin B chain C]. | |||||
|
CTRL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.105315 (rank : 34) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P40313 | Gene names | CTRL, CTRL1 | |||
|
Domain Architecture |
|
|||||
| Description | Chymotrypsin-like protease CTRL-1 precursor (EC 3.4.21.-). | |||||
|
DESC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.102263 (rank : 57) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZ10, Q8BZ04 | Gene names | Desc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease DESC4 precursor (EC 3.4.21.-) [Contains: Serine protease DESC4 non-catalytic chain; Serine protease DESC4 catalytic chain]. | |||||
|
EGFB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.086423 (rank : 151) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P36368, P00754 | Gene names | Egfbp2, Egfbp-2, Klk-13, Klk13 | |||
|
Domain Architecture |
|
|||||
| Description | Epidermal growth factor-binding protein type B precursor (EC 3.4.21.35) (EGF-BP B) (Glandular kallikrein K13) (Tissue kallikrein 13) (mGK-13) (Prorenin-converting enzyme) (PRECE). | |||||
|
ELA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.107862 (rank : 24) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNI1, Q5MLF0, Q6DJT0, Q6ISM6 | Gene names | ELA1 | |||
|
Domain Architecture |
|
|||||
| Description | Elastase-1 precursor (EC 3.4.21.36). | |||||
|
ELA2A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.105878 (rank : 32) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08217, Q14243 | Gene names | ELA2A | |||
|
Domain Architecture |
|
|||||
| Description | Elastase-2A precursor (EC 3.4.21.71). | |||||
|
ELA2A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.107599 (rank : 26) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05208 | Gene names | Ela2a, Ela-2, Ela2 | |||
|
Domain Architecture |
|
|||||
| Description | Elastase-2A precursor (EC 3.4.21.71) (Elastase-2). | |||||
|
ELA2B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.105990 (rank : 30) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08218 | Gene names | ELA2B | |||
|
Domain Architecture |
|
|||||
| Description | Elastase-2B precursor (EC 3.4.21.71). | |||||
|
ELA3A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.109802 (rank : 20) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09093, Q9BRW4 | Gene names | ELA3A, ELA3 | |||
|
Domain Architecture |
|
|||||
| Description | Elastase-3A precursor (EC 3.4.21.70) (Elastase IIIA) (Protease E). | |||||
|
ELA3B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.110224 (rank : 19) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08861, P11423, Q5VU28, Q5VU29, Q5VU30 | Gene names | ELA3B | |||
|
Domain Architecture |
|
|||||
| Description | Elastase-3B precursor (EC 3.4.21.70) (Elastase IIIB) (Protease E). | |||||
|
ELNE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.108865 (rank : 23) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08246, P09649, Q6B0D9, Q6LDP5 | Gene names | ELA2 | |||
|
Domain Architecture |
|
|||||
| Description | Leukocyte elastase precursor (EC 3.4.21.37) (Elastase-2) (Neutrophil elastase) (PMN elastase) (Bone marrow serine protease) (Medullasin) (Human leukocyte elastase) (HLE). | |||||
|
ELNE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.109514 (rank : 22) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UP87, Q61515 | Gene names | Ela2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte elastase precursor (EC 3.4.21.37) (Elastase-2) (Neutrophil elastase). | |||||
|
ENTK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.095022 (rank : 95) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98073 | Gene names | PRSS7, ENTK | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase precursor (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
ENTK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.098441 (rank : 79) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97435 | Gene names | Prss7, Entk | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
FA10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.087999 (rank : 143) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00742, Q14340 | Gene names | F10 | |||
|
Domain Architecture |
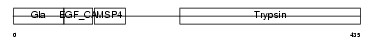
|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) (Stuart- Prower factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
FA10_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.085088 (rank : 159) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88947, O54740, Q99L32 | Gene names | F10 | |||
|
Domain Architecture |
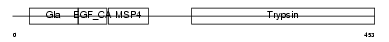
|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
FA11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.096652 (rank : 89) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P03951, Q9Y495 | Gene names | F11 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XI precursor (EC 3.4.21.27) (Plasma thromboplastin antecedent) (PTA) (FXI) [Contains: Coagulation factor XIa heavy chain; Coagulation factor XIa light chain]. | |||||
|
FA11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.095550 (rank : 92) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91Y47 | Gene names | F11 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XI precursor (EC 3.4.21.27) (Plasma thromboplastin antecedent) (PTA) (FXI) [Contains: Coagulation factor XIa heavy chain; Coagulation factor XIa light chain]. | |||||
|
FA12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.107759 (rank : 25) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00748, P78339 | Gene names | F12 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XII precursor (EC 3.4.21.38) (Hageman factor) (HAF) [Contains: Coagulation factor XIIa heavy chain; Beta-factor XIIa part 1; Beta-factor XIIa part 2; Coagulation factor XIIa light chain]. | |||||
|
FA7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.094956 (rank : 96) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08709, Q14339, Q5JVF2 | Gene names | F7 | |||
|
Domain Architecture |
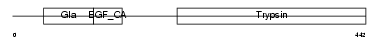
|
|||||
| Description | Coagulation factor VII precursor (EC 3.4.21.21) (Serum prothrombin conversion accelerator) (SPCA) (Proconvertin) (Eptacog alfa) [Contains: Factor VII light chain; Factor VII heavy chain]. | |||||
|
FA7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.091895 (rank : 113) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70375, Q61109 | Gene names | F7, Cf7 | |||
|
Domain Architecture |
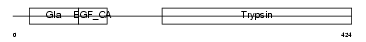
|
|||||
| Description | Coagulation factor VII precursor (EC 3.4.21.21) (Serum prothrombin conversion accelerator) [Contains: Factor VII light chain; Factor VII heavy chain]. | |||||
|
FA9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.085314 (rank : 157) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00740 | Gene names | F9 | |||
|
Domain Architecture |
|
|||||
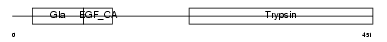
| Description | Coagulation factor IX precursor (EC 3.4.21.22) (Christmas factor) (Plasma thromboplastin component) (PTC) [Contains: Coagulation factor IXa light chain; Coagulation factor IXa heavy chain]. | |||||
|
GRAC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.089639 (rank : 133) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08882, Q61389 | Gene names | Gzmc, Ctla-5, Ctla5 | |||
|
Domain Architecture |
|
|||||
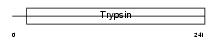
| Description | Granzyme C precursor (EC 3.4.21.-) (Cytotoxic cell protease 2) (CCP2) (B10). | |||||
|
GRAD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.093846 (rank : 103) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11033, P97387 | Gene names | Gzmd | |||
|
Domain Architecture |
|
|||||
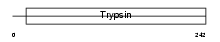
| Description | Granzyme D precursor (EC 3.4.21.-). | |||||
|
GRAE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.097075 (rank : 86) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08884, P97389 | Gene names | Gzme, Ccp3, Ctla-6, Ctla6 | |||
|
Domain Architecture |
|
|||||
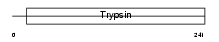
| Description | Granzyme E precursor (EC 3.4.21.-) (Cytotoxic cell protease 3) (CCP3) (CTL serine protease 2) (D12) (Cytotoxic serine protease 2) (MCSP2). | |||||
|
GRAF_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.097906 (rank : 82) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08883 | Gene names | Gzmf, Ccp4, Ctla-7, Ctla7 | |||
|
Domain Architecture |
|
|||||
| Description | Granzyme F precursor (EC 3.4.21.-) (Cytotoxic cell protease 4) (CCP4) (CTL serine protease 3) (C134) (Cytotoxic serine protease 3) (MCSP3). | |||||
|
GRAG_MOUSE
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.097275 (rank : 85) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13366, P97388 | Gene names | Gzmg | |||
|
Domain Architecture |

|
|||||
| Description | Granzyme G precursor (EC 3.4.21.-) (CTL serine protease 1) (MCSP-1). | |||||
|
GRAH_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.093620 (rank : 104) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20718 | Gene names | GZMH, CGL2, CTSGL2 | |||
|
Domain Architecture |
|
|||||
| Description | Granzyme H precursor (EC 3.4.21.-) (Cytotoxic T-lymphocyte proteinase) (Cathepsin G-like 2) (CTSGL2) (CCP-X) (Cytotoxic serine protease C) (CSP-C). | |||||
|
GRAK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.101607 (rank : 59) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35205 | Gene names | Gzmk | |||
|
Domain Architecture |
|
|||||
| Description | Granzyme K precursor (EC 3.4.21.-). | |||||
|
GRAM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.099204 (rank : 72) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51124 | Gene names | GZMM, MET1 | |||
|
Domain Architecture |
|
|||||
| Description | Granzyme M precursor (EC 3.4.21.-) (Met-ase) (Natural killer cell granular protease) (HU-Met-1) (Met-1 serine protease). | |||||
|
HABP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.104532 (rank : 44) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14520, O00663 | Gene names | HABP2, HGFAL, PHBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hyaluronan-binding protein 2 precursor (EC 3.4.21.-) (Plasma hyaluronan-binding protein) (Hepatocyte growth factor activator-like protein) (Factor VII-activating protease) (Factor seven-activating protease) (FSAP) [Contains: Hyaluronan-binding protein 2 50 kDa heavy chain; Hyaluronan-binding protein 2 50 kDa heavy chain alternate form; Hyaluronan-binding protein 2 27 kDa light chain; Hyaluronan-binding protein 2 27 kDa light chain alternate form]. | |||||
|
HABP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.104893 (rank : 39) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K0D2 | Gene names | Habp2, Phbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hyaluronan-binding protein 2 precursor (EC 3.4.21.-) (Plasma hyaluronan-binding protein) [Contains: Hyaluronan-binding protein 2 50 kDa heavy chain; Hyaluronan-binding protein 2 50 kDa heavy chain alternate form; Hyaluronan-binding protein 2 27 kDa light chain; Hyaluronan-binding protein 2 27 kDa light chain alternate form]. | |||||
|
HEPS_HUMAN
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.107208 (rank : 27) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05981 | Gene names | HPN, TMPRSS1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease hepsin (EC 3.4.21.-) (Transmembrane protease, serine 1) [Contains: Serine protease hepsin non-catalytic chain; Serine protease hepsin catalytic chain]. | |||||
|
HEPS_MOUSE
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.105045 (rank : 38) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35453, Q9CW97 | Gene names | Hpn | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease hepsin (EC 3.4.21.-) [Contains: Serine protease hepsin non-catalytic chain; Serine protease hepsin catalytic chain]. | |||||
|
HGFA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.106654 (rank : 29) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
HGFA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.102900 (rank : 52) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R098, Q9JKV4 | Gene names | Hgfac | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
HGFL_HUMAN
|
||||||
| θ value | θ > 10 (rank : 68) | NC score | 0.190518 (rank : 7) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26927, Q13350, Q14870 | Gene names | MST1, HGFL | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor-like protein precursor (Macrophage stimulatory protein) (MSP) (Macrophage-stimulating protein) [Contains: Hepatocyte growth factor-like protein alpha chain; Hepatocyte growth factor-like protein beta chain]. | |||||
|
HGFL_MOUSE
|
||||||
| θ value | θ > 10 (rank : 69) | NC score | 0.188897 (rank : 8) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26928 | Gene names | Mst1, Hgfl | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor-like protein precursor (Macrophage stimulatory protein) (MSP) [Contains: Hepatocyte growth factor-like protein alpha chain; Hepatocyte growth factor-like protein beta chain]. | |||||
|
HPTR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 70) | NC score | 0.103337 (rank : 51) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00739, Q7LE20, Q92658, Q92659, Q9ULB0 | Gene names | HPR | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin-related protein precursor. | |||||
|
HPT_HUMAN
|
||||||
| θ value | θ > 10 (rank : 71) | NC score | 0.099173 (rank : 73) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00738, P00737, Q2PP15, Q3B7J0, Q6LBY9 | Gene names | HP | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin precursor [Contains: Haptoglobin alpha chain; Haptoglobin beta chain]. | |||||
|
HPT_MOUSE
|
||||||
| θ value | θ > 10 (rank : 72) | NC score | 0.109583 (rank : 21) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61646 | Gene names | Hp | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin precursor [Contains: Haptoglobin alpha chain; Haptoglobin beta chain]. | |||||
|
K1B11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 73) | NC score | 0.089581 (rank : 135) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15946 | Gene names | Klk1b11, Klk-11, Klk11 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b11 precursor (EC 3.4.21.35) (Tissue kallikrein-11) (Glandular kallikrein K11) (mGK-11). | |||||
|
K1B16_MOUSE
|
||||||
| θ value | θ > 10 (rank : 74) | NC score | 0.080071 (rank : 170) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04071 | Gene names | Klk1b16, Klk-16, Klk16 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b16 precursor (EC 3.4.21.54) (Gamma- renin, submandibular gland) (mGK-16). | |||||
|
K1B21_MOUSE
|
||||||
| θ value | θ > 10 (rank : 75) | NC score | 0.089049 (rank : 138) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61759, Q61760, Q9JM70 | Gene names | Klk1b21, Klk-21, Klk21 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b21 precursor (EC 3.4.21.35) (Tissue kallikrein 21) (Glandular kallikrein K21) (mGK-21). | |||||
|
K1B22_MOUSE
|
||||||
| θ value | θ > 10 (rank : 76) | NC score | 0.089796 (rank : 130) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15948 | Gene names | Klk1b22, Klk-22, Klk22 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b22 precursor (EC 3.4.21.35) (Glandular kallikrein K22) (Tissue kallikrein 22) (mGK-22) (Epidermal growth factor-binding protein type A) (EGF-BP A) (Nerve growth factor beta chain endopeptidase) (Beta-NGF-endopeptidase). | |||||
|
K1B24_MOUSE
|
||||||
| θ value | θ > 10 (rank : 77) | NC score | 0.089581 (rank : 134) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61754, Q61755, Q9JM69 | Gene names | Klk1b24, Klk-24, Klk24 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein 1-related peptidase b24 precursor (EC 3.4.21.35) (Glandular kallikrein K24) (Tissue kallikrein 24) (mGK-24). | |||||
|
K1B26_MOUSE
|
||||||
| θ value | θ > 10 (rank : 78) | NC score | 0.086004 (rank : 155) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P36369, P00753, P00754 | Gene names | Klk1b26, Klk-26, Klk26 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein 1-related peptidase b26 precursor (EC 3.4.21.35) (Glandular kallikrein K26) (Tissue kallikrein 26) (mGK-26) (Prorenin-converting enzyme 2) (PRECE-2). | |||||
|
K1B27_MOUSE
|
||||||
| θ value | θ > 10 (rank : 79) | NC score | 0.088406 (rank : 141) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JM71 | Gene names | Klk1b27, Klk-27, Klk27 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein 1-related peptidase b27 precursor (EC 3.4.21.-) (Tissue kallikrein 27) (Glandular kallikrein K27) (mGK-27) (mKlk27). | |||||
|
K1KB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 80) | NC score | 0.086234 (rank : 153) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00755 | Gene names | Klk1b1, Klk-1, Klk1 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein 1-related peptidase b1 precursor (EC 3.4.21.35) (Glandular kallikrein K1) (Tissue kallikrein-1) (mGK-1). | |||||
|
K1KB3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 81) | NC score | 0.089720 (rank : 131) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00756 | Gene names | Klk1b3, Klk-3, Klk3, Ngfg | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein 1-related peptidase b3 precursor (EC 3.4.21.35) (Glandular kallikrein K3) (Tissue kallikrein-3) (mGK-3) (7S nerve growth factor gamma chain) (Gamma-NGF) [Contains: Nerve growth factor gamma chain 1; Nerve growth factor gamma chain 2]. | |||||
|
K1KB4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 82) | NC score | 0.086626 (rank : 149) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00757 | Gene names | Klk1b4, Klk-4, Klk4, Ngfa | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein 1-related peptidase-like b4 precursor (7S nerve growth factor alpha chain) (Alpha-NGF). | |||||
|
K1KB5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 83) | NC score | 0.086184 (rank : 154) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15945, Q52KM1, Q565D7 | Gene names | Klk1b5, Klk-5, Klk5 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein 1-related peptidase b5 precursor (EC 3.4.21.35) (Glandular kallikrein K5) (Tissue kallikrein-5) (mGK-5). | |||||
|
K1KB8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 84) | NC score | 0.087904 (rank : 144) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07628 | Gene names | Klk1b8, Klk-8, Klk8 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein 1-related peptidase b8 precursor (EC 3.4.21.35) (Tissue kallikrein-8) (Glandular kallikrein K8) (mGK-8). | |||||
|
K1KB9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 85) | NC score | 0.089900 (rank : 128) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15949 | Gene names | Klk1b9, Egfbp3, Klk-9, Klk9 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein 1-related peptidase b9 precursor (EC 3.4.21.35) (Tissue kallikrein-9) (Glandular kallikrein K9) (mGK-9) (Epidermal growth factor-binding protein type C) (EGF-BP C). | |||||
|
KLK10_HUMAN
|
||||||
| θ value | θ > 10 (rank : 86) | NC score | 0.086787 (rank : 147) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43240, Q99920, Q9GZW9 | Gene names | KLK10, NES1, PRRSL1 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-10 precursor (EC 3.4.21.-) (Protease serine-like 1) (Normal epithelial cell-specific 1). | |||||
|
KLK11_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.093415 (rank : 105) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBX7, O75837, Q9NS65 | Gene names | KLK11, PRSS20, TLSP | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-11 precursor (EC 3.4.21.-) (Hippostasin) (Trypsin-like protease). | |||||
|
KLK11_MOUSE
|
||||||
| θ value | θ > 10 (rank : 88) | NC score | 0.095224 (rank : 94) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYN3, Q9QYN4 | Gene names | Klk11, Prss20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kallikrein-11 precursor (EC 3.4.21.-) (Hippostasin). | |||||
|
KLK12_HUMAN
|
||||||
| θ value | θ > 10 (rank : 89) | NC score | 0.089345 (rank : 136) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKR0, Q9UKR1 | Gene names | KLK12, KLKL5 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-12 precursor (EC 3.4.21.-) (Kallikrein-like protein 5) (KLK-L5). | |||||
|
KLK13_HUMAN
|
||||||
| θ value | θ > 10 (rank : 90) | NC score | 0.090745 (rank : 123) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKR3, Q9Y433 | Gene names | KLK13, KLKL4 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-13 precursor (EC 3.4.21.-) (Kallikrein-like protein 4) (KLK-L4). | |||||
|
KLK14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 91) | NC score | 0.084302 (rank : 164) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0G3 | Gene names | KLK14, KLKL6 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-14 precursor (EC 3.4.21.-) (Kallikrein-like protein 6) (KLK-L6). | |||||
|
KLK15_HUMAN
|
||||||
| θ value | θ > 10 (rank : 92) | NC score | 0.090746 (rank : 122) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H2R5, Q15358, Q9H2R3, Q9H2R4, Q9H2R6, Q9HBG9 | Gene names | KLK15 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-15 precursor (EC 3.4.21.-) (ACO protease). | |||||
|
KLK1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 93) | NC score | 0.091178 (rank : 119) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06870, Q8TCV8, Q9BS53, Q9NQU4, Q9UMJ1 | Gene names | KLK1 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-1 precursor (EC 3.4.21.35) (Tissue kallikrein) (Kidney/pancreas/salivary gland kallikrein). | |||||
|
KLK1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 94) | NC score | 0.086427 (rank : 150) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15947, Q61855 | Gene names | Klk1, Klk-6, Klk6 | |||
|
Domain Architecture |
|
|||||
| Description | Glandular kallikrein K1 precursor (EC 3.4.21.35) (Tissue kallikrein-6) (mGK-6) (Renal kallikrein) (KAL-B). | |||||
|
KLK2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 95) | NC score | 0.092139 (rank : 112) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20151, Q15946, Q9UJZ9 | Gene names | KLK2 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-2 precursor (EC 3.4.21.35) (Tissue kallikrein-2) (Glandular kallikrein-1) (hGK-1). | |||||
|
KLK3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 96) | NC score | 0.092666 (rank : 109) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07288, Q16272, Q86TG8 | Gene names | KLK3, APS | |||
|
Domain Architecture |
|
|||||
| Description | Prostate-specific antigen precursor (EC 3.4.21.77) (PSA) (Kallikrein- 3) (Semenogelase) (Gamma-seminoprotein) (Seminin) (P-30 antigen). | |||||
|
KLK4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 97) | NC score | 0.083248 (rank : 166) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5K2, Q4VB16, Q9GZL6, Q9UBJ6 | Gene names | KLK4, EMSP1, PRSS17, PSTS | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-4 precursor (EC 3.4.21.-) (Prostase) (Kallikrein-like protein 1) (KLK-L1) (Enamel matrix serine proteinase 1). | |||||
|
KLK5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 98) | NC score | 0.088611 (rank : 140) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y337, Q9HBG8 | Gene names | KLK5, SCTE | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-5 precursor (EC 3.4.21.-) (Stratum corneum tryptic enzyme) (Kallikrein-like protein 2) (KLK-L2). | |||||
|
KLK6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 99) | NC score | 0.091194 (rank : 118) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92876 | Gene names | KLK6, PRSS18, PRSS9 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-6 precursor (EC 3.4.21.-) (Protease M) (Neurosin) (Zyme) (SP59). | |||||
|
KLK7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 100) | NC score | 0.082886 (rank : 168) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49862, Q8N5N9, Q8NFV7 | Gene names | KLK7, PRSS6, SCCE | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-7 precursor (EC 3.4.21.-) (hK7) (Stratum corneum chymotryptic enzyme) (hSCCE). | |||||
|
KLK7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 101) | NC score | 0.084617 (rank : 163) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91VE3, Q9R048 | Gene names | Klk7, Prss6, Scce | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kallikrein-7 precursor (EC 3.4.21.-) (Stratum corneum chymotryptic enzyme) (Thymopsin). | |||||
|
KLK9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 102) | NC score | 0.088801 (rank : 139) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKQ9 | Gene names | KLK9 | |||
|
Domain Architecture |
|
|||||
| Description | Kallikrein-9 precursor (EC 3.4.21.-) (Kallikrein-like protein 3) (KLK- L3). | |||||
|
KLKB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 103) | NC score | 0.096298 (rank : 91) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P03952 | Gene names | KLKB1, KLK3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma kallikrein precursor (EC 3.4.21.34) (Plasma prekallikrein) (Kininogenin) (Fletcher factor) [Contains: Plasma kallikrein heavy chain; Plasma kallikrein light chain]. | |||||
|
KLKB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 104) | NC score | 0.098573 (rank : 78) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26262 | Gene names | Klkb1, Klk3, Pk | |||
|
Domain Architecture |

|
|||||
| Description | Plasma kallikrein precursor (EC 3.4.21.34) (Plasma prekallikrein) (Kininogenin) (Fletcher factor) [Contains: Plasma kallikrein heavy chain; Plasma kallikrein light chain]. | |||||
|
KREM1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 105) | NC score | 0.141629 (rank : 10) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96MU8, Q5TIB9, Q6P3X6, Q9BY70, Q9UGS5, Q9UGU1 | Gene names | KREMEN1, KREMEN, KRM1 | |||
|
Domain Architecture |
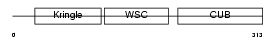
|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
KREM1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 106) | NC score | 0.142444 (rank : 9) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99N43 | Gene names | Kremen1, Kremen | |||
|
Domain Architecture |
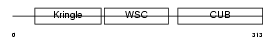
|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
KREM2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 107) | NC score | 0.133120 (rank : 12) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NCW0, Q8N2J4, Q8NCW1, Q96GL8, Q9BTP9 | Gene names | KREMEN2, KRM2 | |||
|
Domain Architecture |
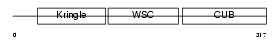
|
|||||
| Description | Kremen protein 2 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 2) (Dickkopf receptor 2). | |||||
|
KREM2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 108) | NC score | 0.135624 (rank : 11) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1S7 | Gene names | Kremen2 | |||
|
Domain Architecture |
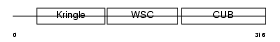
|
|||||
| Description | Kremen protein 2 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 2) (Dickkopf receptor 2). | |||||
|
MASP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 109) | NC score | 0.083587 (rank : 165) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
MASP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 110) | NC score | 0.084892 (rank : 162) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
MASP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 111) | NC score | 0.082035 (rank : 169) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
MASP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 112) | NC score | 0.083175 (rank : 167) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
MCPT1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 113) | NC score | 0.091549 (rank : 116) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23946, Q16018 | Gene names | CMA1, CYH, CYM | |||
|
Domain Architecture |
|
|||||
| Description | Chymase precursor (EC 3.4.21.39) (Mast cell protease I). | |||||
|
MCPT1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 114) | NC score | 0.090448 (rank : 126) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11034 | Gene names | Mcpt1 | |||
|
Domain Architecture |
|
|||||
| Description | Mast cell protease 1 precursor (EC 3.4.21.-) (MMCP-1). | |||||
|
MCPT2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 115) | NC score | 0.090856 (rank : 120) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15119 | Gene names | Mcpt2 | |||
|
Domain Architecture |
|
|||||
| Description | Mast cell protease 2 precursor (EC 3.4.21.-) (MMCP-2). | |||||
|
MCPT4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 116) | NC score | 0.090630 (rank : 124) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21812, Q9EPQ9, Q9EQT2 | Gene names | Mcpt4 | |||
|
Domain Architecture |
|
|||||
| Description | Mast cell protease 4 precursor (EC 3.4.21.-) (MMCP-4) (Serosal mast cell protease) (MSMCP) (Myonase). | |||||
|
MCPT5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 117) | NC score | 0.092673 (rank : 108) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21844, Q9R1F0 | Gene names | Mcpt5 | |||
|
Domain Architecture |
|
|||||
| Description | Mast cell protease 5 precursor (EC 3.4.21.-) (MMCP-5) (Mast cell chymase 1). | |||||
|
MCPT6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 118) | NC score | 0.100981 (rank : 65) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21845, Q61962 | Gene names | Mcpt6 | |||
|
Domain Architecture |
|
|||||
| Description | Mast cell protease 6 precursor (EC 3.4.21.59) (MMCP-6) (Tryptase). | |||||
|
MCPT8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 119) | NC score | 0.086948 (rank : 146) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43430, Q9R1F1 | Gene names | Mcpt8 | |||
|
Domain Architecture |
|
|||||
| Description | Mast cell protease 8 precursor (EC 3.4.21.-) (MMCP-8). | |||||
|
MCPT9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 120) | NC score | 0.096894 (rank : 87) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35164 | Gene names | Mcpt9 | |||
|
Domain Architecture |
|
|||||
| Description | Mast cell protease 9 precursor (EC 3.4.21.-) (MMCP-9). | |||||
|
MCPTX_MOUSE
|
||||||
| θ value | θ > 10 (rank : 121) | NC score | 0.098638 (rank : 77) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00356 | Gene names | Mcptl | |||
|
Domain Architecture |
|
|||||
| Description | Mast cell protease-like protein precursor (EC 3.4.21.-). | |||||
|
NETR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 122) | NC score | 0.090509 (rank : 125) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56730, Q9UP16 | Gene names | PRSS12 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Leydin). | |||||
|
NETR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 123) | NC score | 0.091605 (rank : 115) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08762 | Gene names | Prss12, Bssp3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Brain-specific serine protease 3) (BSSP-3). | |||||
|
NRPN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 124) | NC score | 0.086349 (rank : 152) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60259, Q9HCB3, Q9UIL9, Q9UQ47 | Gene names | KLK8, NRPN, PRSS19, TADG14 | |||
|
Domain Architecture |
|
|||||
| Description | Neuropsin precursor (EC 3.4.21.-) (NP) (Kallikrein-8) (Ovasin) (Serine protease TADG-14) (Tumor-associated differentially expressed gene 14 protein). | |||||
|
NRPN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 125) | NC score | 0.089833 (rank : 129) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61955 | Gene names | Klk8, Nrpn, Prss19 | |||
|
Domain Architecture |
|
|||||
| Description | Neuropsin precursor (EC 3.4.21.-) (NP) (Kallikrein-8). | |||||
|
POLS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 126) | NC score | 0.099988 (rank : 68) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5K4E3, Q8NBY4 | Gene names | PRSS36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyserase-2 precursor (EC 3.4.21.-) (Polyserine protease 2) (Protease serine 36). | |||||
|
POLS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 127) | NC score | 0.099634 (rank : 69) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5K2P8 | Gene names | Prss36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyserase-2 precursor (EC 3.4.21.-) (Polyserine protease 2) (Protease serine 36). | |||||
|
PROC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 128) | NC score | 0.090338 (rank : 127) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04070, Q15189, Q15190, Q16001 | Gene names | PROC | |||
|
Domain Architecture |
|
|||||
| Description | Vitamin K-dependent protein C precursor (EC 3.4.21.69) (Autoprothrombin IIA) (Anticoagulant protein C) (Blood coagulation factor XIV) [Contains: Vitamin K-dependent protein C light chain; Vitamin K-dependent protein C heavy chain; Activation peptide]. | |||||
|
PROC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 129) | NC score | 0.087036 (rank : 145) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 4 | |
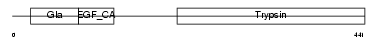
| SwissProt Accessions | P33587, O35498, Q91WN8, Q99PC6 | Gene names | Proc | |||
|
Domain Architecture |
|
|||||
| Description | Vitamin K-dependent protein C precursor (EC 3.4.21.69) (Autoprothrombin IIA) (Anticoagulant protein C) (Blood coagulation factor XIV) [Contains: Vitamin K-dependent protein C light chain; Vitamin K-dependent protein C heavy chain; Activation peptide]. | |||||
|
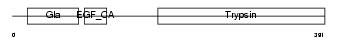
PROZ_HUMAN
|
||||||
| θ value | θ > 10 (rank : 130) | NC score | 0.055069 (rank : 178) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22891, Q15213 | Gene names | PROZ | |||
|
Domain Architecture |
|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
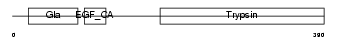
PROZ_MOUSE
|
||||||
| θ value | θ > 10 (rank : 131) | NC score | 0.053031 (rank : 179) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQW3 | Gene names | Proz | |||
|
Domain Architecture |
|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
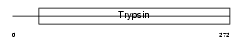
PRS27_HUMAN
|
||||||
| θ value | θ > 10 (rank : 132) | NC score | 0.104765 (rank : 41) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQR3 | Gene names | PRSS27, MPN | |||
|
Domain Architecture |
|
|||||
| Description | Protease serine 27 precursor (EC 3.4.21.-) (Marapsin) (Pancreasin) (Channel-activating protease 2) (CAPH2). | |||||
|
PRS27_MOUSE
|
||||||
| θ value | θ > 10 (rank : 133) | NC score | 0.104469 (rank : 45) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BJR6 | Gene names | Prss27, Mpn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protease serine 27 precursor (EC 3.4.21.-) (Marapsin) (Pancreasin). | |||||
|
PRSS8_HUMAN
|
||||||
| θ value | θ > 10 (rank : 134) | NC score | 0.098051 (rank : 81) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16651, Q9UCA3 | Gene names | PRSS8 | |||
|
Domain Architecture |
|
|||||
| Description | Prostasin precursor (EC 3.4.21.-) [Contains: Prostasin light chain; Prostasin heavy chain]. | |||||
|
PRSS8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 135) | NC score | 0.099467 (rank : 71) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ESD1 | Gene names | Prss8, Cap1 | |||
|
Domain Architecture |
|
|||||
| Description | Prostasin precursor (EC 3.4.21.-) (Channel-activating protease 1) [Contains: Prostasin light chain; Prostasin heavy chain]. | |||||
|
PRTN3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 136) | NC score | 0.102180 (rank : 58) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24158, P15637, P18078, Q6LBM7, Q9UQD8 | Gene names | PRTN3, MBN | |||
|
Domain Architecture |
|
|||||
| Description | Myeloblastin precursor (EC 3.4.21.76) (Leukocyte proteinase 3) (PR-3) (PR3) (AGP7) (Wegener autoantigen) (P29) (C-ANCA antigen) (Neutrophil proteinase 4) (NP-4). | |||||
|
PRTN3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 137) | NC score | 0.106658 (rank : 28) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61096, O08809 | Gene names | Prtn3 | |||
|
Domain Architecture |
|
|||||
| Description | Myeloblastin precursor (EC 3.4.21.76) (Proteinase 3) (PR-3). | |||||
|
ST14_HUMAN
|
||||||
| θ value | θ > 10 (rank : 138) | NC score | 0.089647 (rank : 132) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5Y6, Q9BS01, Q9H3S0, Q9HB36, Q9HCA3 | Gene names | ST14, PRSS14, SNC19, TADG15 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Matriptase) (Membrane-type serine protease 1) (MT-SP1) (Prostamin) (Serine protease TADG-15) (Tumor-associated differentially-expressed gene 15 protein). | |||||
|
ST14_MOUSE
|
||||||
| θ value | θ > 10 (rank : 139) | NC score | 0.088362 (rank : 142) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56677 | Gene names | St14, Prss14 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Epithin). | |||||
|
TEST_HUMAN
|
||||||
| θ value | θ > 10 (rank : 140) | NC score | 0.104426 (rank : 47) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6M0, Q9NS34, Q9P2V6 | Gene names | PRSS21, ESP1, TEST1 | |||
|
Domain Architecture |
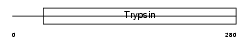
|
|||||
| Description | Testisin precursor (EC 3.4.21.-) (Eosinophil serine protease 1) (ESP- 1). | |||||
|
TEST_MOUSE
|
||||||
| θ value | θ > 10 (rank : 141) | NC score | 0.105099 (rank : 36) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHJ7, Q9DA14 | Gene names | Prss21 | |||
|
Domain Architecture |
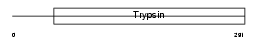
|
|||||
| Description | Testisin precursor (EC 3.4.21.-) (Tryptase 4). | |||||
|
THRB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 142) | NC score | 0.130359 (rank : 14) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00734 | Gene names | F2 | |||
|
Domain Architecture |

|
|||||
| Description | Prothrombin precursor (EC 3.4.21.5) (Coagulation factor II) [Contains: Activation peptide fragment 1; Activation peptide fragment 2; Thrombin light chain; Thrombin heavy chain]. | |||||
|
THRB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 143) | NC score | 0.130777 (rank : 13) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19221 | Gene names | F2, Cf2 | |||
|
Domain Architecture |

|
|||||
| Description | Prothrombin precursor (EC 3.4.21.5) (Coagulation factor II) [Contains: Activation peptide fragment 1; Activation peptide fragment 2; Thrombin light chain; Thrombin heavy chain]. | |||||
|
TM11D_HUMAN
|
||||||
| θ value | θ > 10 (rank : 144) | NC score | 0.097332 (rank : 84) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60235 | Gene names | TMPRSS11D, HAT | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 11D precursor (EC 3.4.21.-) (Airway trypsin-like protease) [Contains: Transmembrane protease, serine 11D non-catalytic chain; Transmembrane protease, serine 11D catalytic chain]. | |||||
|
TM11D_MOUSE
|
||||||
| θ value | θ > 10 (rank : 145) | NC score | 0.097441 (rank : 83) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHK8, Q7TNX3, Q8VDV1 | Gene names | Tmprss11d, Mat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 11D precursor (EC 3.4.21.-) (Airway trypsin-like protease) (AT) (Adrenal secretory serine protease) (AsP) [Contains: Transmembrane protease, serine 11D non-catalytic chain; Transmembrane protease, serine 11D catalytic chain]. | |||||
|
TM11E_HUMAN
|
||||||
| θ value | θ > 10 (rank : 146) | NC score | 0.098975 (rank : 74) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UL52, Q6UW31 | Gene names | TMPRSS11E, DESC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 11E precursor (EC 3.4.21.-) (Serine protease DESC1) [Contains: Transmembrane protease, serine 11E non- catalytic chain; Transmembrane protease, serine 11E catalytic chain]. | |||||
|
TM11E_MOUSE
|
||||||
| θ value | θ > 10 (rank : 147) | NC score | 0.100746 (rank : 66) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5S248, Q8BM10 | Gene names | Tmprss11e, Desc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 11E precursor (EC 3.4.21.-) (Serine protease DESC1) [Contains: Transmembrane protease, serine 11E non- catalytic chain; Transmembrane protease, serine 11E catalytic chain]. | |||||
|
TMPS2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 148) | NC score | 0.101402 (rank : 63) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15393, Q9BXX1 | Gene names | TMPRSS2, PRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 precursor (EC 3.4.21.-) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
TMPS2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 149) | NC score | 0.104432 (rank : 46) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 4 | |
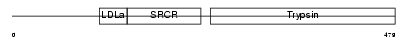
| SwissProt Accessions | Q9JIQ8, Q9JKC4, Q9QY82 | Gene names | Tmprss2 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 2 (EC 3.4.21.-) (Epitheliasin) (Plasmic transmembrane protein X) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
TMPS3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 150) | NC score | 0.103890 (rank : 50) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57727, Q6ZMC3 | Gene names | TMPRSS3, ECHOS1, TADG12 | |||
|
Domain Architecture |
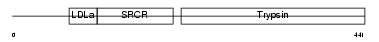
|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-) (Serine protease TADG- 12) (Tumor-associated differentially-expressed gene 12 protein). | |||||
|
TMPS3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 151) | NC score | 0.104591 (rank : 43) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1T0, Q8VDE0 | Gene names | Tmprss3 | |||
|
Domain Architecture |
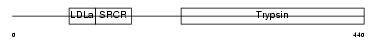
|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-). | |||||
|
TMPS4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 152) | NC score | 0.101488 (rank : 60) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRS4, Q5XKQ6, Q6UX37, Q9NZA5 | Gene names | TMPRSS4, TMPRSS3 | |||
|
Domain Architecture |
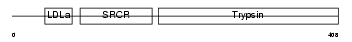
|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Membrane-type serine protease 2) (MT-SP2). | |||||
|
TMPS4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 153) | NC score | 0.104074 (rank : 49) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCA5 | Gene names | Tmprss4, Cap2 | |||
|
Domain Architecture |
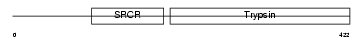
|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Channel-activating protease 2) (mCAP2). | |||||
|
TMPS5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 154) | NC score | 0.098052 (rank : 80) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3S3 | Gene names | TMPRSS5 | |||
|
Domain Architecture |
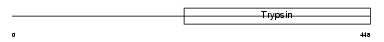
|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
TMPS5_MOUSE
|
||||||
| θ value | θ > 10 (rank : 155) | NC score | 0.096673 (rank : 88) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ER04, Q9ER02, Q9ER03 | Gene names | Tmprss5 | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
TMPS6_HUMAN
|
||||||
| θ value | θ > 10 (rank : 156) | NC score | 0.090812 (rank : 121) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IU80, Q5TI06, Q6UXD8, Q8IUE2, Q8IXV8 | Gene names | TMPRSS6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
TMPS6_MOUSE
|
||||||
| θ value | θ > 10 (rank : 157) | NC score | 0.093261 (rank : 107) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBI0, Q6PF94 | Gene names | Tmprss6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
TMPS7_HUMAN
|
||||||
| θ value | θ > 10 (rank : 158) | NC score | 0.091834 (rank : 114) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7RTY8 | Gene names | TMPRSS7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
TMPS7_MOUSE
|
||||||
| θ value | θ > 10 (rank : 159) | NC score | 0.092232 (rank : 110) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BIK6 | Gene names | Tmprss7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
TMPS9_HUMAN
|
||||||
| θ value | θ > 10 (rank : 160) | NC score | 0.094500 (rank : 100) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z410, Q6ZND6, Q7Z411 | Gene names | TMPRSS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 9 (EC 3.4.21.-) (Polyserase-1) (Polyserase-I) (Polyserine protease 1) [Contains: Serase-1; Serase-2; Serase-3]. | |||||
|
TMPS9_MOUSE
|
||||||
| θ value | θ > 10 (rank : 161) | NC score | 0.093298 (rank : 106) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P69525 | Gene names | Tmprss9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 9 (EC 3.4.21.-) (Polyserase-1) (Polyserine protease 1) (Polyserase-I) [Contains: Serase-1; Serase-2; Serase-3]. | |||||
|
TMPSD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 162) | NC score | 0.102848 (rank : 53) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
TMPSD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 163) | NC score | 0.102297 (rank : 56) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
TMSP8_MOUSE
|
||||||
| θ value | θ > 10 (rank : 164) | NC score | 0.105859 (rank : 33) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYZ9, Q91XC4 | Gene names | Tmprss8, Disp | |||
|
Domain Architecture |
|
|||||
| Description | Transmembrane serine protease 8 precursor (EC 3.4.21.-) (Distal intestinal serine protease). | |||||
|
TPA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 165) | NC score | 0.127760 (rank : 16) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00750, Q15103, Q7Z7N2, Q86YK8, Q9BU99 | Gene names | PLAT | |||
|
Domain Architecture |

|
|||||
| Description | Tissue-type plasminogen activator precursor (EC 3.4.21.68) (tPA) (t- PA) (t-plasminogen activator) (Alteplase) (Reteplase) [Contains: Tissue-type plasminogen activator chain A; Tissue-type plasminogen activator chain B]. | |||||
|
TPA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 166) | NC score | 0.128149 (rank : 15) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11214, Q91VP2 | Gene names | Plat | |||
|
Domain Architecture |

|
|||||
| Description | Tissue-type plasminogen activator precursor (EC 3.4.21.68) (tPA) (t- PA) (t-plasminogen activator) [Contains: Tissue-type plasminogen activator chain A; Tissue-type plasminogen activator chain B]. | |||||
|
TRY1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 167) | NC score | 0.094464 (rank : 101) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07477, Q92955, Q9HAN4, Q9HAN5, Q9HAN6, Q9HAN7 | Gene names | PRSS1, TRP1, TRY1, TRYP1 | |||
|
Domain Architecture |
|
|||||
| Description | Trypsin-1 precursor (EC 3.4.21.4) (Trypsin I) (Cationic trypsinogen). | |||||
|
TRY2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 168) | NC score | 0.094647 (rank : 98) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07478 | Gene names | PRSS2, TRY2, TRYP2 | |||
|
Domain Architecture |
|
|||||
| Description | Trypsin-2 precursor (EC 3.4.21.4) (Trypsin II) (Anionic trypsinogen). | |||||
|
TRY2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 169) | NC score | 0.094565 (rank : 99) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07146 | Gene names | Prss2, Try2 | |||
|
Domain Architecture |
|
|||||
| Description | Anionic trypsin-2 precursor (EC 3.4.21.4) (Anionic trypsin II) (Pretrypsinogen II). | |||||
|
TRY3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 170) | NC score | 0.095316 (rank : 93) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35030, P15951, Q15665, Q9UQV3 | Gene names | PRSS3, PRSS4, TRY3, TRY4 | |||
|
Domain Architecture |
|
|||||
| Description | Trypsin-3 precursor (EC 3.4.21.4) (Trypsin III) (Brain trypsinogen) (Mesotrypsinogen) (Trypsin IV). | |||||
|
TRYA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 171) | NC score | 0.100607 (rank : 67) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15157, Q9H2Y5, Q9UQI1 | Gene names | TPSAB1, TPS1 | |||
|
Domain Architecture |
|
|||||
| Description | Tryptase alpha-1 precursor (EC 3.4.21.59) (Tryptase-1). | |||||
|
TRYB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 172) | NC score | 0.098759 (rank : 75) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15661, Q15663, Q9H2Y4 | Gene names | TPSAB1, TPSB1 | |||
|
Domain Architecture |
|
|||||
| Description | Tryptase beta-1 precursor (EC 3.4.21.59) (Tryptase-1) (Tryptase I). | |||||
|
TRYB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 173) | NC score | 0.105260 (rank : 35) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02844 | Gene names | Tpsab1, Mcpt7 | |||
|
Domain Architecture |

|
|||||
| Description | Tryptase precursor (EC 3.4.21.59) (Mast cell protease 7) (MMCP-7) (Tryptase alpha/beta-1). | |||||
|
TRYB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 174) | NC score | 0.098759 (rank : 76) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20231, O95827, Q15664, Q9UQI6, Q9UQI7 | Gene names | TPSB2, TPS2 | |||
|
Domain Architecture |
|
|||||
| Description | Tryptase beta-2 precursor (EC 3.4.21.59) (Tryptase-2) (Tryptase II). | |||||
|
TRYD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 175) | NC score | 0.101441 (rank : 62) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZJ3, O95824, Q8TDI6, Q96L36, Q96RZ5, Q9H2Y6, Q9UQI8 | Gene names | TPSD1 | |||
|
Domain Architecture |
|
|||||
| Description | Tryptase delta precursor (EC 3.4.21.59) (Delta tryptase) (Mast cell mMCP-7-like) (Tryptase-3) (HmMCP-3-like tryptase III). | |||||
|
TRYG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 176) | NC score | 0.105886 (rank : 31) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRR2, Q96RZ8, Q9C015, Q9NRQ8, Q9UBB2 | Gene names | TPSG1, TMT | |||
|
Domain Architecture |
|
|||||
| Description | Tryptase gamma precursor (EC 3.4.21.-) (Transmembrane tryptase) [Contains: Tryptase gamma light chain; Tryptase gamma heavy chain]. | |||||
|
TRYG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 177) | NC score | 0.102707 (rank : 55) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QUL7 | Gene names | Tpsg1, Tmt | |||
|
Domain Architecture |
|
|||||
| Description | Tryptase gamma precursor (EC 3.4.21.-) (Transmembrane tryptase) [Contains: Tryptase gamma light chain; Tryptase gamma heavy chain]. | |||||
|
TSP50_HUMAN
|
||||||
| θ value | θ > 10 (rank : 178) | NC score | 0.104776 (rank : 40) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UI38 | Gene names | TSP50 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protease-like protein 50 precursor. | |||||
|
UROK_HUMAN
|
||||||
| θ value | θ > 10 (rank : 179) | NC score | 0.111059 (rank : 18) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00749, Q15844, Q16618, Q969W6 | Gene names | PLAU | |||
|
Domain Architecture |

|
|||||
| Description | Urokinase-type plasminogen activator precursor (EC 3.4.21.73) (uPA) (U-plasminogen activator) [Contains: Urokinase-type plasminogen activator long chain A; Urokinase-type plasminogen activator short chain A; Urokinase-type plasminogen activator chain B]. | |||||
|
UROK_MOUSE
|
||||||
| θ value | θ > 10 (rank : 180) | NC score | 0.111709 (rank : 17) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06869 | Gene names | Plau | |||
|
Domain Architecture |

|
|||||
| Description | Urokinase-type plasminogen activator precursor (EC 3.4.21.73) (uPA) (U-plasminogen activator) [Contains: Urokinase-type plasminogen activator long chain A; Urokinase-type plasminogen activator short chain A; Urokinase-type plasminogen activator chain B]. | |||||
|
PRGB_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 5.96599e-36 (rank : 1) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q02325 | Gene names | PLGL, PRGB | |||
|
Domain Architecture |

|
|||||
| Description | Plasminogen-related protein B precursor. | |||||
|
PLMN_HUMAN
|
||||||
| NC score | 0.246510 (rank : 2) | θ value | 1.24399e-33 (rank : 2) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 204 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P00747, Q15146, Q6PA00 | Gene names | PLG | |||
|
Domain Architecture |

|
|||||
| Description | Plasminogen precursor (EC 3.4.21.7) [Contains: Plasmin heavy chain A; Activation peptide; Angiostatin; Plasmin heavy chain A, short form; Plasmin light chain B]. | |||||
|
PLMN_MOUSE
|
||||||
| NC score | 0.244021 (rank : 3) | θ value | 7.54701e-31 (rank : 3) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20918, Q8CIS2, Q91WJ5 | Gene names | Plg | |||
|
Domain Architecture |

|
|||||
| Description | Plasminogen precursor (EC 3.4.21.7) [Contains: Plasmin heavy chain A; Activation peptide; Angiostatin; Plasmin heavy chain A, short form; Plasmin light chain B]. | |||||
|
HGF_HUMAN
|
||||||
| NC score | 0.199178 (rank : 4) | θ value | 0.62314 (rank : 4) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 191 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P14210, Q02935, Q13494, Q14519, Q8TCE2, Q9BYL9, Q9BYM0, Q9UDU6 | Gene names | HGF, HPTA | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor precursor (Scatter factor) (SF) (Hepatopoeitin A) [Contains: Hepatocyte growth factor alpha chain; Hepatocyte growth factor beta chain]. | |||||
|
HGF_MOUSE
|
||||||
| NC score | 0.196779 (rank : 5) | θ value | 4.03905 (rank : 6) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q08048, Q61662, Q64007, Q6LBE6 | Gene names | Hgf | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor precursor (Scatter factor) (SF) (Hepatopoeitin A) [Contains: Hepatocyte growth factor alpha chain; Hepatocyte growth factor beta chain]. | |||||
|
APOA_HUMAN
|
||||||
| NC score | 0.190519 (rank : 6) | θ value | θ > 10 (rank : 9) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08519 | Gene names | LPA | |||
|
Domain Architecture |

|
|||||
| Description | Apolipoprotein(a) precursor (EC 3.4.21.-) (Apo(a)) (Lp(a)). | |||||
|
HGFL_HUMAN
|
||||||
| NC score | 0.190518 (rank : 7) | θ value | θ > 10 (rank : 68) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 190 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26927, Q13350, Q14870 | Gene names | MST1, HGFL | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor-like protein precursor (Macrophage stimulatory protein) (MSP) (Macrophage-stimulating protein) [Contains: Hepatocyte growth factor-like protein alpha chain; Hepatocyte growth factor-like protein beta chain]. | |||||
|
HGFL_MOUSE
|
||||||
| NC score | 0.188897 (rank : 8) | θ value | θ > 10 (rank : 69) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26928 | Gene names | Mst1, Hgfl | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor-like protein precursor (Macrophage stimulatory protein) (MSP) [Contains: Hepatocyte growth factor-like protein alpha chain; Hepatocyte growth factor-like protein beta chain]. | |||||
|
KREM1_MOUSE
|
||||||
| NC score | 0.142444 (rank : 9) | θ value | θ > 10 (rank : 106) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 92 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99N43 | Gene names | Kremen1, Kremen | |||
|
Domain Architecture |

|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
KREM1_HUMAN
|
||||||
| NC score | 0.141629 (rank : 10) | θ value | θ > 10 (rank : 105) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96MU8, Q5TIB9, Q6P3X6, Q9BY70, Q9UGS5, Q9UGU1 | Gene names | KREMEN1, KREMEN, KRM1 | |||
|
Domain Architecture |

|
|||||
| Description | Kremen protein 1 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 1) (Dickkopf receptor). | |||||
|
KREM2_MOUSE
|
||||||
| NC score | 0.135624 (rank : 11) | θ value | θ > 10 (rank : 108) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 90 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1S7 | Gene names | Kremen2 | |||
|
Domain Architecture |

|
|||||
| Description | Kremen protein 2 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 2) (Dickkopf receptor 2). | |||||
|
KREM2_HUMAN
|
||||||
| NC score | 0.133120 (rank : 12) | θ value | θ > 10 (rank : 107) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 93 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8NCW0, Q8N2J4, Q8NCW1, Q96GL8, Q9BTP9 | Gene names | KREMEN2, KRM2 | |||
|
Domain Architecture |

|
|||||
| Description | Kremen protein 2 precursor (Kringle-containing protein marking the eye and the nose) (Kringle domain-containing transmembrane protein 2) (Dickkopf receptor 2). | |||||
|
THRB_MOUSE
|
||||||
| NC score | 0.130777 (rank : 13) | θ value | θ > 10 (rank : 143) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P19221 | Gene names | F2, Cf2 | |||
|
Domain Architecture |

|
|||||
| Description | Prothrombin precursor (EC 3.4.21.5) (Coagulation factor II) [Contains: Activation peptide fragment 1; Activation peptide fragment 2; Thrombin light chain; Thrombin heavy chain]. | |||||
|
THRB_HUMAN
|
||||||
| NC score | 0.130359 (rank : 14) | θ value | θ > 10 (rank : 142) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00734 | Gene names | F2 | |||
|
Domain Architecture |

|
|||||
| Description | Prothrombin precursor (EC 3.4.21.5) (Coagulation factor II) [Contains: Activation peptide fragment 1; Activation peptide fragment 2; Thrombin light chain; Thrombin heavy chain]. | |||||
|
TPA_MOUSE
|
||||||
| NC score | 0.128149 (rank : 15) | θ value | θ > 10 (rank : 166) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 335 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11214, Q91VP2 | Gene names | Plat | |||
|
Domain Architecture |

|
|||||
| Description | Tissue-type plasminogen activator precursor (EC 3.4.21.68) (tPA) (t- PA) (t-plasminogen activator) [Contains: Tissue-type plasminogen activator chain A; Tissue-type plasminogen activator chain B]. | |||||
|
TPA_HUMAN
|
||||||
| NC score | 0.127760 (rank : 16) | θ value | θ > 10 (rank : 165) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00750, Q15103, Q7Z7N2, Q86YK8, Q9BU99 | Gene names | PLAT | |||
|
Domain Architecture |

|
|||||
| Description | Tissue-type plasminogen activator precursor (EC 3.4.21.68) (tPA) (t- PA) (t-plasminogen activator) (Alteplase) (Reteplase) [Contains: Tissue-type plasminogen activator chain A; Tissue-type plasminogen activator chain B]. | |||||
|
UROK_MOUSE
|
||||||
| NC score | 0.111709 (rank : 17) | θ value | θ > 10 (rank : 180) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 273 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06869 | Gene names | Plau | |||
|
Domain Architecture |

|
|||||
| Description | Urokinase-type plasminogen activator precursor (EC 3.4.21.73) (uPA) (U-plasminogen activator) [Contains: Urokinase-type plasminogen activator long chain A; Urokinase-type plasminogen activator short chain A; Urokinase-type plasminogen activator chain B]. | |||||
|
UROK_HUMAN
|
||||||
| NC score | 0.111059 (rank : 18) | θ value | θ > 10 (rank : 179) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 272 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00749, Q15844, Q16618, Q969W6 | Gene names | PLAU | |||
|
Domain Architecture |

|
|||||
| Description | Urokinase-type plasminogen activator precursor (EC 3.4.21.73) (uPA) (U-plasminogen activator) [Contains: Urokinase-type plasminogen activator long chain A; Urokinase-type plasminogen activator short chain A; Urokinase-type plasminogen activator chain B]. | |||||
|
ELA3B_HUMAN
|
||||||
| NC score | 0.110224 (rank : 19) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08861, P11423, Q5VU28, Q5VU29, Q5VU30 | Gene names | ELA3B | |||
|
Domain Architecture |

|
|||||
| Description | Elastase-3B precursor (EC 3.4.21.70) (Elastase IIIB) (Protease E). | |||||
|
ELA3A_HUMAN
|
||||||
| NC score | 0.109802 (rank : 20) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09093, Q9BRW4 | Gene names | ELA3A, ELA3 | |||
|
Domain Architecture |

|
|||||
| Description | Elastase-3A precursor (EC 3.4.21.70) (Elastase IIIA) (Protease E). | |||||
|
HPT_MOUSE
|
||||||
| NC score | 0.109583 (rank : 21) | θ value | θ > 10 (rank : 72) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61646 | Gene names | Hp | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin precursor [Contains: Haptoglobin alpha chain; Haptoglobin beta chain]. | |||||
|
ELNE_MOUSE
|
||||||
| NC score | 0.109514 (rank : 22) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q3UP87, Q61515 | Gene names | Ela2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leukocyte elastase precursor (EC 3.4.21.37) (Elastase-2) (Neutrophil elastase). | |||||
|
ELNE_HUMAN
|
||||||
| NC score | 0.108865 (rank : 23) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 164 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08246, P09649, Q6B0D9, Q6LDP5 | Gene names | ELA2 | |||
|
Domain Architecture |

|
|||||
| Description | Leukocyte elastase precursor (EC 3.4.21.37) (Elastase-2) (Neutrophil elastase) (PMN elastase) (Bone marrow serine protease) (Medullasin) (Human leukocyte elastase) (HLE). | |||||
|
ELA1_HUMAN
|
||||||
| NC score | 0.107862 (rank : 24) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UNI1, Q5MLF0, Q6DJT0, Q6ISM6 | Gene names | ELA1 | |||
|
Domain Architecture |

|
|||||
| Description | Elastase-1 precursor (EC 3.4.21.36). | |||||
|
FA12_HUMAN
|
||||||
| NC score | 0.107759 (rank : 25) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 425 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00748, P78339 | Gene names | F12 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XII precursor (EC 3.4.21.38) (Hageman factor) (HAF) [Contains: Coagulation factor XIIa heavy chain; Beta-factor XIIa part 1; Beta-factor XIIa part 2; Coagulation factor XIIa light chain]. | |||||
|
ELA2A_MOUSE
|
||||||
| NC score | 0.107599 (rank : 26) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05208 | Gene names | Ela2a, Ela-2, Ela2 | |||
|
Domain Architecture |

|
|||||
| Description | Elastase-2A precursor (EC 3.4.21.71) (Elastase-2). | |||||
|
HEPS_HUMAN
|
||||||
| NC score | 0.107208 (rank : 27) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05981 | Gene names | HPN, TMPRSS1 | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease hepsin (EC 3.4.21.-) (Transmembrane protease, serine 1) [Contains: Serine protease hepsin non-catalytic chain; Serine protease hepsin catalytic chain]. | |||||
|
PRTN3_MOUSE
|
||||||
| NC score | 0.106658 (rank : 28) | θ value | θ > 10 (rank : 137) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61096, O08809 | Gene names | Prtn3 | |||
|
Domain Architecture |

|
|||||
| Description | Myeloblastin precursor (EC 3.4.21.76) (Proteinase 3) (PR-3). | |||||
|
HGFA_HUMAN
|
||||||
| NC score | 0.106654 (rank : 29) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 436 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q04756, Q14726 | Gene names | HGFAC | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
ELA2B_HUMAN
|
||||||
| NC score | 0.105990 (rank : 30) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08218 | Gene names | ELA2B | |||
|
Domain Architecture |

|
|||||
| Description | Elastase-2B precursor (EC 3.4.21.71). | |||||
|
TRYG1_HUMAN
|
||||||
| NC score | 0.105886 (rank : 31) | θ value | θ > 10 (rank : 176) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRR2, Q96RZ8, Q9C015, Q9NRQ8, Q9UBB2 | Gene names | TPSG1, TMT | |||
|
Domain Architecture |

|
|||||
| Description | Tryptase gamma precursor (EC 3.4.21.-) (Transmembrane tryptase) [Contains: Tryptase gamma light chain; Tryptase gamma heavy chain]. | |||||
|
ELA2A_HUMAN
|
||||||
| NC score | 0.105878 (rank : 32) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08217, Q14243 | Gene names | ELA2A | |||
|
Domain Architecture |

|
|||||
| Description | Elastase-2A precursor (EC 3.4.21.71). | |||||
|
TMSP8_MOUSE
|
||||||
| NC score | 0.105859 (rank : 33) | θ value | θ > 10 (rank : 164) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYZ9, Q91XC4 | Gene names | Tmprss8, Disp | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane serine protease 8 precursor (EC 3.4.21.-) (Distal intestinal serine protease). | |||||
|
CTRL_HUMAN
|
||||||
| NC score | 0.105315 (rank : 34) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P40313 | Gene names | CTRL, CTRL1 | |||
|
Domain Architecture |

|
|||||
| Description | Chymotrypsin-like protease CTRL-1 precursor (EC 3.4.21.-). | |||||
|
TRYB1_MOUSE
|
||||||
| NC score | 0.105260 (rank : 35) | θ value | θ > 10 (rank : 173) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q02844 | Gene names | Tpsab1, Mcpt7 | |||
|
Domain Architecture |

|
|||||
| Description | Tryptase precursor (EC 3.4.21.59) (Mast cell protease 7) (MMCP-7) (Tryptase alpha/beta-1). | |||||
|
TEST_MOUSE
|
||||||
| NC score | 0.105099 (rank : 36) | θ value | θ > 10 (rank : 141) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JHJ7, Q9DA14 | Gene names | Prss21 | |||
|
Domain Architecture |

|
|||||
| Description | Testisin precursor (EC 3.4.21.-) (Tryptase 4). | |||||
|
CLCR_HUMAN
|
||||||
| NC score | 0.105086 (rank : 37) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99895, O00765, Q9NUH5 | Gene names | CTRC, CLCR | |||
|
Domain Architecture |

|
|||||
| Description | Caldecrin precursor (EC 3.4.21.2) (Chymotrypsin C). | |||||
|
HEPS_MOUSE
|
||||||
| NC score | 0.105045 (rank : 38) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35453, Q9CW97 | Gene names | Hpn | |||
|
Domain Architecture |

|
|||||
| Description | Serine protease hepsin (EC 3.4.21.-) [Contains: Serine protease hepsin non-catalytic chain; Serine protease hepsin catalytic chain]. | |||||
|
HABP2_MOUSE
|
||||||
| NC score | 0.104893 (rank : 39) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K0D2 | Gene names | Habp2, Phbp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hyaluronan-binding protein 2 precursor (EC 3.4.21.-) (Plasma hyaluronan-binding protein) [Contains: Hyaluronan-binding protein 2 50 kDa heavy chain; Hyaluronan-binding protein 2 50 kDa heavy chain alternate form; Hyaluronan-binding protein 2 27 kDa light chain; Hyaluronan-binding protein 2 27 kDa light chain alternate form]. | |||||
|
TSP50_HUMAN
|
||||||
| NC score | 0.104776 (rank : 40) | θ value | θ > 10 (rank : 178) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UI38 | Gene names | TSP50 | |||
|
Domain Architecture |

|
|||||
| Description | Testis-specific protease-like protein 50 precursor. | |||||
|
PRS27_HUMAN
|
||||||
| NC score | 0.104765 (rank : 41) | θ value | θ > 10 (rank : 132) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BQR3 | Gene names | PRSS27, MPN | |||
|
Domain Architecture |

|
|||||
| Description | Protease serine 27 precursor (EC 3.4.21.-) (Marapsin) (Pancreasin) (Channel-activating protease 2) (CAPH2). | |||||
|
ACRO_MOUSE
|
||||||
| NC score | 0.104653 (rank : 42) | θ value | θ > 10 (rank : 8) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23578 | Gene names | Acr | |||
|
Domain Architecture |

|
|||||
| Description | Acrosin precursor (EC 3.4.21.10) [Contains: Acrosin light chain; Acrosin heavy chain]. | |||||
|
TMPS3_MOUSE
|
||||||
| NC score | 0.104591 (rank : 43) | θ value | θ > 10 (rank : 151) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 239 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8K1T0, Q8VDE0 | Gene names | Tmprss3 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-). | |||||
|
HABP2_HUMAN
|
||||||
| NC score | 0.104532 (rank : 44) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q14520, O00663 | Gene names | HABP2, HGFAL, PHBP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hyaluronan-binding protein 2 precursor (EC 3.4.21.-) (Plasma hyaluronan-binding protein) (Hepatocyte growth factor activator-like protein) (Factor VII-activating protease) (Factor seven-activating protease) (FSAP) [Contains: Hyaluronan-binding protein 2 50 kDa heavy chain; Hyaluronan-binding protein 2 50 kDa heavy chain alternate form; Hyaluronan-binding protein 2 27 kDa light chain; Hyaluronan-binding protein 2 27 kDa light chain alternate form]. | |||||
|
PRS27_MOUSE
|
||||||
| NC score | 0.104469 (rank : 45) | θ value | θ > 10 (rank : 133) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BJR6 | Gene names | Prss27, Mpn | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protease serine 27 precursor (EC 3.4.21.-) (Marapsin) (Pancreasin). | |||||
|
TMPS2_MOUSE
|
||||||
| NC score | 0.104432 (rank : 46) | θ value | θ > 10 (rank : 149) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JIQ8, Q9JKC4, Q9QY82 | Gene names | Tmprss2 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 (EC 3.4.21.-) (Epitheliasin) (Plasmic transmembrane protein X) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
TEST_HUMAN
|
||||||
| NC score | 0.104426 (rank : 47) | θ value | θ > 10 (rank : 140) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y6M0, Q9NS34, Q9P2V6 | Gene names | PRSS21, ESP1, TEST1 | |||
|
Domain Architecture |

|
|||||
| Description | Testisin precursor (EC 3.4.21.-) (Eosinophil serine protease 1) (ESP- 1). | |||||
|
CTRB1_HUMAN
|
||||||
| NC score | 0.104254 (rank : 48) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P17538 | Gene names | CTRB1, CTRB | |||
|
Domain Architecture |

|
|||||
| Description | Chymotrypsinogen B precursor (EC 3.4.21.1) [Contains: Chymotrypsin B chain A; Chymotrypsin B chain B; Chymotrypsin B chain C]. | |||||
|
TMPS4_MOUSE
|
||||||
| NC score | 0.104074 (rank : 49) | θ value | θ > 10 (rank : 153) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VCA5 | Gene names | Tmprss4, Cap2 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Channel-activating protease 2) (mCAP2). | |||||
|
TMPS3_HUMAN
|
||||||
| NC score | 0.103890 (rank : 50) | θ value | θ > 10 (rank : 150) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 245 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P57727, Q6ZMC3 | Gene names | TMPRSS3, ECHOS1, TADG12 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 3 (EC 3.4.21.-) (Serine protease TADG- 12) (Tumor-associated differentially-expressed gene 12 protein). | |||||
|
HPTR_HUMAN
|
||||||
| NC score | 0.103337 (rank : 51) | θ value | θ > 10 (rank : 70) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 166 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00739, Q7LE20, Q92658, Q92659, Q9ULB0 | Gene names | HPR | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin-related protein precursor. | |||||
|
HGFA_MOUSE
|
||||||
| NC score | 0.102900 (rank : 52) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9R098, Q9JKV4 | Gene names | Hgfac | |||
|
Domain Architecture |

|
|||||
| Description | Hepatocyte growth factor activator precursor (EC 3.4.21.-) (HGF activator) (HGFA) [Contains: Hepatocyte growth factor activator short chain; Hepatocyte growth factor activator long chain]. | |||||
|
TMPSD_HUMAN
|
||||||
| NC score | 0.102848 (rank : 53) | θ value | θ > 10 (rank : 162) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 333 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BYE2, Q86YM4, Q96JY8, Q9BYE1 | Gene names | TMPRSS13, MSP, TMPRSS11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
ACRO_HUMAN
|
||||||
| NC score | 0.102725 (rank : 54) | θ value | θ > 10 (rank : 7) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 291 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P10323, Q6ICK2 | Gene names | ACR, ACRS | |||
|
Domain Architecture |

|
|||||
| Description | Acrosin precursor (EC 3.4.21.10) [Contains: Acrosin light chain; Acrosin heavy chain]. | |||||
|
TRYG1_MOUSE
|
||||||
| NC score | 0.102707 (rank : 55) | θ value | θ > 10 (rank : 177) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QUL7 | Gene names | Tpsg1, Tmt | |||
|
Domain Architecture |

|
|||||
| Description | Tryptase gamma precursor (EC 3.4.21.-) (Transmembrane tryptase) [Contains: Tryptase gamma light chain; Tryptase gamma heavy chain]. | |||||
|
TMPSD_MOUSE
|
||||||
| NC score | 0.102297 (rank : 56) | θ value | θ > 10 (rank : 163) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 334 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5U405, Q8CFE0, Q91VQ8 | Gene names | Tmprss13, Msp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 13 (EC 3.4.21.-) (Mosaic serine protease) (Membrane-type mosaic serine protease). | |||||
|
DESC4_MOUSE
|
||||||
| NC score | 0.102263 (rank : 57) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BZ10, Q8BZ04 | Gene names | Desc4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine protease DESC4 precursor (EC 3.4.21.-) [Contains: Serine protease DESC4 non-catalytic chain; Serine protease DESC4 catalytic chain]. | |||||
|
PRTN3_HUMAN
|
||||||
| NC score | 0.102180 (rank : 58) | θ value | θ > 10 (rank : 136) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24158, P15637, P18078, Q6LBM7, Q9UQD8 | Gene names | PRTN3, MBN | |||
|
Domain Architecture |

|
|||||
| Description | Myeloblastin precursor (EC 3.4.21.76) (Leukocyte proteinase 3) (PR-3) (PR3) (AGP7) (Wegener autoantigen) (P29) (C-ANCA antigen) (Neutrophil proteinase 4) (NP-4). | |||||
|
GRAK_MOUSE
|
||||||
| NC score | 0.101607 (rank : 59) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35205 | Gene names | Gzmk | |||
|
Domain Architecture |

|
|||||
| Description | Granzyme K precursor (EC 3.4.21.-). | |||||
|
TMPS4_HUMAN
|
||||||
| NC score | 0.101488 (rank : 60) | θ value | θ > 10 (rank : 152) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9NRS4, Q5XKQ6, Q6UX37, Q9NZA5 | Gene names | TMPRSS4, TMPRSS3 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 4 (EC 3.4.21.-) (Membrane-type serine protease 2) (MT-SP2). | |||||
|
BSSP4_HUMAN
|
||||||
| NC score | 0.101466 (rank : 61) | θ value | θ > 10 (rank : 10) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9GZN4, O43342, Q6UXE0 | Gene names | PRSS22, BSSP4, PRSS26 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific serine protease 4 precursor (EC 3.4.21.-) (BSSP-4) (Serine protease 22) (Tryptase epsilon). | |||||
|
TRYD_HUMAN
|
||||||
| NC score | 0.101441 (rank : 62) | θ value | θ > 10 (rank : 175) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9BZJ3, O95824, Q8TDI6, Q96L36, Q96RZ5, Q9H2Y6, Q9UQI8 | Gene names | TPSD1 | |||
|
Domain Architecture |

|
|||||
| Description | Tryptase delta precursor (EC 3.4.21.59) (Delta tryptase) (Mast cell mMCP-7-like) (Tryptase-3) (HmMCP-3-like tryptase III). | |||||
|
TMPS2_HUMAN
|
||||||
| NC score | 0.101402 (rank : 63) | θ value | θ > 10 (rank : 148) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O15393, Q9BXX1 | Gene names | TMPRSS2, PRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 precursor (EC 3.4.21.-) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
CAP7_HUMAN
|
||||||
| NC score | 0.101030 (rank : 64) | θ value | θ > 10 (rank : 16) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20160, P80014, Q52LG4 | Gene names | AZU1 | |||
|
Domain Architecture |

|
|||||
| Description | Azurocidin precursor (Cationic antimicrobial protein CAP37) (Heparin- binding protein) (HBP). | |||||
|
MCPT6_MOUSE
|
||||||
| NC score | 0.100981 (rank : 65) | θ value | θ > 10 (rank : 118) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21845, Q61962 | Gene names | Mcpt6 | |||
|
Domain Architecture |

|
|||||
| Description | Mast cell protease 6 precursor (EC 3.4.21.59) (MMCP-6) (Tryptase). | |||||
|
TM11E_MOUSE
|
||||||
| NC score | 0.100746 (rank : 66) | θ value | θ > 10 (rank : 147) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5S248, Q8BM10 | Gene names | Tmprss11e, Desc1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 11E precursor (EC 3.4.21.-) (Serine protease DESC1) [Contains: Transmembrane protease, serine 11E non- catalytic chain; Transmembrane protease, serine 11E catalytic chain]. | |||||
|
TRYA1_HUMAN
|
||||||
| NC score | 0.100607 (rank : 67) | θ value | θ > 10 (rank : 171) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15157, Q9H2Y5, Q9UQI1 | Gene names | TPSAB1, TPS1 | |||
|
Domain Architecture |

|
|||||
| Description | Tryptase alpha-1 precursor (EC 3.4.21.59) (Tryptase-1). | |||||
|
POLS2_HUMAN
|
||||||
| NC score | 0.099988 (rank : 68) | θ value | θ > 10 (rank : 126) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 192 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5K4E3, Q8NBY4 | Gene names | PRSS36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyserase-2 precursor (EC 3.4.21.-) (Polyserine protease 2) (Protease serine 36). | |||||
|
POLS2_MOUSE
|
||||||
| NC score | 0.099634 (rank : 69) | θ value | θ > 10 (rank : 127) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 188 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5K2P8 | Gene names | Prss36 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Polyserase-2 precursor (EC 3.4.21.-) (Polyserine protease 2) (Protease serine 36). | |||||
|
BSSP4_MOUSE
|
||||||
| NC score | 0.099605 (rank : 70) | θ value | θ > 10 (rank : 11) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ER10 | Gene names | Prss22, Bssp4, Prss26 | |||
|
Domain Architecture |

|
|||||
| Description | Brain-specific serine protease 4 precursor (EC 3.4.21.-) (BSSP-4) (Serine protease 22) (Tryptase epsilon). | |||||
|
PRSS8_MOUSE
|
||||||
| NC score | 0.099467 (rank : 71) | θ value | θ > 10 (rank : 135) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ESD1 | Gene names | Prss8, Cap1 | |||
|
Domain Architecture |

|
|||||
| Description | Prostasin precursor (EC 3.4.21.-) (Channel-activating protease 1) [Contains: Prostasin light chain; Prostasin heavy chain]. | |||||
|
GRAM_HUMAN
|
||||||
| NC score | 0.099204 (rank : 72) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P51124 | Gene names | GZMM, MET1 | |||
|
Domain Architecture |

|
|||||
| Description | Granzyme M precursor (EC 3.4.21.-) (Met-ase) (Natural killer cell granular protease) (HU-Met-1) (Met-1 serine protease). | |||||
|
HPT_HUMAN
|
||||||
| NC score | 0.099173 (rank : 73) | θ value | θ > 10 (rank : 71) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00738, P00737, Q2PP15, Q3B7J0, Q6LBY9 | Gene names | HP | |||
|
Domain Architecture |

|
|||||
| Description | Haptoglobin precursor [Contains: Haptoglobin alpha chain; Haptoglobin beta chain]. | |||||
|
TM11E_HUMAN
|
||||||
| NC score | 0.098975 (rank : 74) | θ value | θ > 10 (rank : 146) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UL52, Q6UW31 | Gene names | TMPRSS11E, DESC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 11E precursor (EC 3.4.21.-) (Serine protease DESC1) [Contains: Transmembrane protease, serine 11E non- catalytic chain; Transmembrane protease, serine 11E catalytic chain]. | |||||
|
TRYB1_HUMAN
|
||||||
| NC score | 0.098759 (rank : 75) | θ value | θ > 10 (rank : 172) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15661, Q15663, Q9H2Y4 | Gene names | TPSAB1, TPSB1 | |||
|
Domain Architecture |

|
|||||
| Description | Tryptase beta-1 precursor (EC 3.4.21.59) (Tryptase-1) (Tryptase I). | |||||
|
TRYB2_HUMAN
|
||||||
| NC score | 0.098759 (rank : 76) | θ value | θ > 10 (rank : 174) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20231, O95827, Q15664, Q9UQI6, Q9UQI7 | Gene names | TPSB2, TPS2 | |||
|
Domain Architecture |

|
|||||
| Description | Tryptase beta-2 precursor (EC 3.4.21.59) (Tryptase-2) (Tryptase II). | |||||
|
MCPTX_MOUSE
|
||||||
| NC score | 0.098638 (rank : 77) | θ value | θ > 10 (rank : 121) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q00356 | Gene names | Mcptl | |||
|
Domain Architecture |

|
|||||
| Description | Mast cell protease-like protein precursor (EC 3.4.21.-). | |||||
|
KLKB1_MOUSE
|
||||||
| NC score | 0.098573 (rank : 78) | θ value | θ > 10 (rank : 104) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P26262 | Gene names | Klkb1, Klk3, Pk | |||
|
Domain Architecture |

|
|||||
| Description | Plasma kallikrein precursor (EC 3.4.21.34) (Plasma prekallikrein) (Kininogenin) (Fletcher factor) [Contains: Plasma kallikrein heavy chain; Plasma kallikrein light chain]. | |||||
|
ENTK_MOUSE
|
||||||
| NC score | 0.098441 (rank : 79) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P97435 | Gene names | Prss7, Entk | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
TMPS5_HUMAN
|
||||||
| NC score | 0.098052 (rank : 80) | θ value | θ > 10 (rank : 154) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 181 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H3S3 | Gene names | TMPRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
PRSS8_HUMAN
|
||||||
| NC score | 0.098051 (rank : 81) | θ value | θ > 10 (rank : 134) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q16651, Q9UCA3 | Gene names | PRSS8 | |||
|
Domain Architecture |

|
|||||
| Description | Prostasin precursor (EC 3.4.21.-) [Contains: Prostasin light chain; Prostasin heavy chain]. | |||||
|
GRAF_MOUSE
|
||||||
| NC score | 0.097906 (rank : 82) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08883 | Gene names | Gzmf, Ccp4, Ctla-7, Ctla7 | |||
|
Domain Architecture |

|
|||||
| Description | Granzyme F precursor (EC 3.4.21.-) (Cytotoxic cell protease 4) (CCP4) (CTL serine protease 3) (C134) (Cytotoxic serine protease 3) (MCSP3). | |||||
|
TM11D_MOUSE
|
||||||
| NC score | 0.097441 (rank : 83) | θ value | θ > 10 (rank : 145) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VHK8, Q7TNX3, Q8VDV1 | Gene names | Tmprss11d, Mat | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 11D precursor (EC 3.4.21.-) (Airway trypsin-like protease) (AT) (Adrenal secretory serine protease) (AsP) [Contains: Transmembrane protease, serine 11D non-catalytic chain; Transmembrane protease, serine 11D catalytic chain]. | |||||
|
TM11D_HUMAN
|
||||||
| NC score | 0.097332 (rank : 84) | θ value | θ > 10 (rank : 144) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60235 | Gene names | TMPRSS11D, HAT | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 11D precursor (EC 3.4.21.-) (Airway trypsin-like protease) [Contains: Transmembrane protease, serine 11D non-catalytic chain; Transmembrane protease, serine 11D catalytic chain]. | |||||
|
GRAG_MOUSE
|
||||||
| NC score | 0.097275 (rank : 85) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P13366, P97388 | Gene names | Gzmg | |||
|
Domain Architecture |

|
|||||
| Description | Granzyme G precursor (EC 3.4.21.-) (CTL serine protease 1) (MCSP-1). | |||||
|
GRAE_MOUSE
|
||||||
| NC score | 0.097075 (rank : 86) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08884, P97389 | Gene names | Gzme, Ccp3, Ctla-6, Ctla6 | |||
|
Domain Architecture |

|
|||||
| Description | Granzyme E precursor (EC 3.4.21.-) (Cytotoxic cell protease 3) (CCP3) (CTL serine protease 2) (D12) (Cytotoxic serine protease 2) (MCSP2). | |||||
|
MCPT9_MOUSE
|
||||||
| NC score | 0.096894 (rank : 87) | θ value | θ > 10 (rank : 120) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O35164 | Gene names | Mcpt9 | |||
|
Domain Architecture |

|
|||||
| Description | Mast cell protease 9 precursor (EC 3.4.21.-) (MMCP-9). | |||||
|
TMPS5_MOUSE
|
||||||
| NC score | 0.096673 (rank : 88) | θ value | θ > 10 (rank : 155) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 185 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9ER04, Q9ER02, Q9ER03 | Gene names | Tmprss5 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 5 (EC 3.4.21.-) (Spinesin). | |||||
|
FA11_HUMAN
|
||||||
| NC score | 0.096652 (rank : 89) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P03951, Q9Y495 | Gene names | F11 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XI precursor (EC 3.4.21.27) (Plasma thromboplastin antecedent) (PTA) (FXI) [Contains: Coagulation factor XIa heavy chain; Coagulation factor XIa light chain]. | |||||
|
CATG_MOUSE
|
||||||
| NC score | 0.096566 (rank : 90) | θ value | θ > 10 (rank : 18) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P28293 | Gene names | Ctsg | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin G precursor (EC 3.4.21.20) (Vimentin-specific protease) (VSP). | |||||
|
KLKB1_HUMAN
|
||||||
| NC score | 0.096298 (rank : 91) | θ value | θ > 10 (rank : 103) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P03952 | Gene names | KLKB1, KLK3 | |||
|
Domain Architecture |

|
|||||
| Description | Plasma kallikrein precursor (EC 3.4.21.34) (Plasma prekallikrein) (Kininogenin) (Fletcher factor) [Contains: Plasma kallikrein heavy chain; Plasma kallikrein light chain]. | |||||
|
FA11_MOUSE
|
||||||
| NC score | 0.095550 (rank : 92) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91Y47 | Gene names | F11 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor XI precursor (EC 3.4.21.27) (Plasma thromboplastin antecedent) (PTA) (FXI) [Contains: Coagulation factor XIa heavy chain; Coagulation factor XIa light chain]. | |||||
|
TRY3_HUMAN
|
||||||
| NC score | 0.095316 (rank : 93) | θ value | θ > 10 (rank : 170) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P35030, P15951, Q15665, Q9UQV3 | Gene names | PRSS3, PRSS4, TRY3, TRY4 | |||
|
Domain Architecture |

|
|||||
| Description | Trypsin-3 precursor (EC 3.4.21.4) (Trypsin III) (Brain trypsinogen) (Mesotrypsinogen) (Trypsin IV). | |||||
|
KLK11_MOUSE
|
||||||
| NC score | 0.095224 (rank : 94) | θ value | θ > 10 (rank : 88) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9QYN3, Q9QYN4 | Gene names | Klk11, Prss20 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kallikrein-11 precursor (EC 3.4.21.-) (Hippostasin). | |||||
|
ENTK_HUMAN
|
||||||
| NC score | 0.095022 (rank : 95) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 310 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98073 | Gene names | PRSS7, ENTK | |||
|
Domain Architecture |

|
|||||
| Description | Enteropeptidase precursor (EC 3.4.21.9) (Enterokinase) [Contains: Enteropeptidase non-catalytic heavy chain; Enteropeptidase catalytic light chain]. | |||||
|
FA7_HUMAN
|
||||||
| NC score | 0.094956 (rank : 96) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 404 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08709, Q14339, Q5JVF2 | Gene names | F7 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VII precursor (EC 3.4.21.21) (Serum prothrombin conversion accelerator) (SPCA) (Proconvertin) (Eptacog alfa) [Contains: Factor VII light chain; Factor VII heavy chain]. | |||||
|
CFAD_MOUSE
|
||||||
| NC score | 0.094907 (rank : 97) | θ value | θ > 10 (rank : 22) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P03953, Q61280 | Gene names | Cfd, Adn, Df | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor D precursor (EC 3.4.21.46) (C3 convertase activator) (Properdin factor D) (Adipsin) (28 kDa adipocyte protein). | |||||
|
TRY2_HUMAN
|
||||||
| NC score | 0.094647 (rank : 98) | θ value | θ > 10 (rank : 168) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07478 | Gene names | PRSS2, TRY2, TRYP2 | |||
|
Domain Architecture |

|
|||||
| Description | Trypsin-2 precursor (EC 3.4.21.4) (Trypsin II) (Anionic trypsinogen). | |||||
|
TRY2_MOUSE
|
||||||
| NC score | 0.094565 (rank : 99) | θ value | θ > 10 (rank : 169) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07146 | Gene names | Prss2, Try2 | |||
|
Domain Architecture |

|
|||||
| Description | Anionic trypsin-2 precursor (EC 3.4.21.4) (Anionic trypsin II) (Pretrypsinogen II). | |||||
|
TMPS9_HUMAN
|
||||||
| NC score | 0.094500 (rank : 100) | θ value | θ > 10 (rank : 160) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 276 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7Z410, Q6ZND6, Q7Z411 | Gene names | TMPRSS9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 9 (EC 3.4.21.-) (Polyserase-1) (Polyserase-I) (Polyserine protease 1) [Contains: Serase-1; Serase-2; Serase-3]. | |||||
|
TRY1_HUMAN
|
||||||
| NC score | 0.094464 (rank : 101) | θ value | θ > 10 (rank : 167) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07477, Q92955, Q9HAN4, Q9HAN5, Q9HAN6, Q9HAN7 | Gene names | PRSS1, TRP1, TRY1, TRYP1 | |||
|
Domain Architecture |

|
|||||
| Description | Trypsin-1 precursor (EC 3.4.21.4) (Trypsin I) (Cationic trypsinogen). | |||||
|
CFAD_HUMAN
|
||||||
| NC score | 0.094298 (rank : 102) | θ value | θ > 10 (rank : 21) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00746 | Gene names | CFD, DF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor D precursor (EC 3.4.21.46) (C3 convertase activator) (Properdin factor D) (Adipsin). | |||||
|
GRAD_MOUSE
|
||||||
| NC score | 0.093846 (rank : 103) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11033, P97387 | Gene names | Gzmd | |||
|
Domain Architecture |

|
|||||
| Description | Granzyme D precursor (EC 3.4.21.-). | |||||
|
GRAH_HUMAN
|
||||||
| NC score | 0.093620 (rank : 104) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20718 | Gene names | GZMH, CGL2, CTSGL2 | |||
|
Domain Architecture |

|
|||||
| Description | Granzyme H precursor (EC 3.4.21.-) (Cytotoxic T-lymphocyte proteinase) (Cathepsin G-like 2) (CTSGL2) (CCP-X) (Cytotoxic serine protease C) (CSP-C). | |||||
|
KLK11_HUMAN
|
||||||
| NC score | 0.093415 (rank : 105) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UBX7, O75837, Q9NS65 | Gene names | KLK11, PRSS20, TLSP | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-11 precursor (EC 3.4.21.-) (Hippostasin) (Trypsin-like protease). | |||||
|
TMPS9_MOUSE
|
||||||
| NC score | 0.093298 (rank : 106) | θ value | θ > 10 (rank : 161) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 296 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P69525 | Gene names | Tmprss9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 9 (EC 3.4.21.-) (Polyserase-1) (Polyserine protease 1) (Polyserase-I) [Contains: Serase-1; Serase-2; Serase-3]. | |||||
|
TMPS6_MOUSE
|
||||||
| NC score | 0.093261 (rank : 107) | θ value | θ > 10 (rank : 157) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 341 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9DBI0, Q6PF94 | Gene names | Tmprss6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
MCPT5_MOUSE
|
||||||
| NC score | 0.092673 (rank : 108) | θ value | θ > 10 (rank : 117) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21844, Q9R1F0 | Gene names | Mcpt5 | |||
|
Domain Architecture |

|
|||||
| Description | Mast cell protease 5 precursor (EC 3.4.21.-) (MMCP-5) (Mast cell chymase 1). | |||||
|
KLK3_HUMAN
|
||||||
| NC score | 0.092666 (rank : 109) | θ value | θ > 10 (rank : 96) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07288, Q16272, Q86TG8 | Gene names | KLK3, APS | |||
|
Domain Architecture |

|
|||||
| Description | Prostate-specific antigen precursor (EC 3.4.21.77) (PSA) (Kallikrein- 3) (Semenogelase) (Gamma-seminoprotein) (Seminin) (P-30 antigen). | |||||
|
TMPS7_MOUSE
|
||||||
| NC score | 0.092232 (rank : 110) | θ value | θ > 10 (rank : 159) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BIK6 | Gene names | Tmprss7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
CFAI_HUMAN
|
||||||
| NC score | 0.092152 (rank : 111) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 289 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P05156, O60442 | Gene names | CFI, IF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
KLK2_HUMAN
|
||||||
| NC score | 0.092139 (rank : 112) | θ value | θ > 10 (rank : 95) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P20151, Q15946, Q9UJZ9 | Gene names | KLK2 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-2 precursor (EC 3.4.21.35) (Tissue kallikrein-2) (Glandular kallikrein-1) (hGK-1). | |||||
|
FA7_MOUSE
|
||||||
| NC score | 0.091895 (rank : 113) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 374 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70375, Q61109 | Gene names | F7, Cf7 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor VII precursor (EC 3.4.21.21) (Serum prothrombin conversion accelerator) [Contains: Factor VII light chain; Factor VII heavy chain]. | |||||
|
TMPS7_HUMAN
|
||||||
| NC score | 0.091834 (rank : 114) | θ value | θ > 10 (rank : 158) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 275 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q7RTY8 | Gene names | TMPRSS7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Transmembrane protease, serine 7 precursor (EC 3.4.21.-). | |||||
|
NETR_MOUSE
|
||||||
| NC score | 0.091605 (rank : 115) | θ value | θ > 10 (rank : 123) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O08762 | Gene names | Prss12, Bssp3 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Brain-specific serine protease 3) (BSSP-3). | |||||
|
MCPT1_HUMAN
|
||||||
| NC score | 0.091549 (rank : 116) | θ value | θ > 10 (rank : 113) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P23946, Q16018 | Gene names | CMA1, CYH, CYM | |||
|
Domain Architecture |

|
|||||
| Description | Chymase precursor (EC 3.4.21.39) (Mast cell protease I). | |||||
|
CATG_HUMAN
|
||||||
| NC score | 0.091313 (rank : 117) | θ value | θ > 10 (rank : 17) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08311 | Gene names | CTSG | |||
|
Domain Architecture |

|
|||||
| Description | Cathepsin G precursor (EC 3.4.21.20) (CG). | |||||
|
KLK6_HUMAN
|
||||||
| NC score | 0.091194 (rank : 118) | θ value | θ > 10 (rank : 99) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q92876 | Gene names | KLK6, PRSS18, PRSS9 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-6 precursor (EC 3.4.21.-) (Protease M) (Neurosin) (Zyme) (SP59). | |||||
|
KLK1_HUMAN
|
||||||
| NC score | 0.091178 (rank : 119) | θ value | θ > 10 (rank : 93) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06870, Q8TCV8, Q9BS53, Q9NQU4, Q9UMJ1 | Gene names | KLK1 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-1 precursor (EC 3.4.21.35) (Tissue kallikrein) (Kidney/pancreas/salivary gland kallikrein). | |||||
|
MCPT2_MOUSE
|
||||||
| NC score | 0.090856 (rank : 120) | θ value | θ > 10 (rank : 115) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15119 | Gene names | Mcpt2 | |||
|
Domain Architecture |

|
|||||
| Description | Mast cell protease 2 precursor (EC 3.4.21.-) (MMCP-2). | |||||
|
TMPS6_HUMAN
|
||||||
| NC score | 0.090812 (rank : 121) | θ value | θ > 10 (rank : 156) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 338 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8IU80, Q5TI06, Q6UXD8, Q8IUE2, Q8IXV8 | Gene names | TMPRSS6 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 6 (EC 3.4.21.-) (Matriptase-2). | |||||
|
KLK15_HUMAN
|
||||||
| NC score | 0.090746 (rank : 122) | θ value | θ > 10 (rank : 92) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9H2R5, Q15358, Q9H2R3, Q9H2R4, Q9H2R6, Q9HBG9 | Gene names | KLK15 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-15 precursor (EC 3.4.21.-) (ACO protease). | |||||
|
KLK13_HUMAN
|
||||||
| NC score | 0.090745 (rank : 123) | θ value | θ > 10 (rank : 90) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKR3, Q9Y433 | Gene names | KLK13, KLKL4 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-13 precursor (EC 3.4.21.-) (Kallikrein-like protein 4) (KLK-L4). | |||||
|
MCPT4_MOUSE
|
||||||
| NC score | 0.090630 (rank : 124) | θ value | θ > 10 (rank : 116) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21812, Q9EPQ9, Q9EQT2 | Gene names | Mcpt4 | |||
|
Domain Architecture |

|
|||||
| Description | Mast cell protease 4 precursor (EC 3.4.21.-) (MMCP-4) (Serosal mast cell protease) (MSMCP) (Myonase). | |||||
|
NETR_HUMAN
|
||||||
| NC score | 0.090509 (rank : 125) | θ value | θ > 10 (rank : 122) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56730, Q9UP16 | Gene names | PRSS12 | |||
|
Domain Architecture |

|
|||||
| Description | Neurotrypsin precursor (EC 3.4.21.-) (Motopsin) (Leydin). | |||||
|
MCPT1_MOUSE
|
||||||
| NC score | 0.090448 (rank : 126) | θ value | θ > 10 (rank : 114) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P11034 | Gene names | Mcpt1 | |||
|
Domain Architecture |

|
|||||
| Description | Mast cell protease 1 precursor (EC 3.4.21.-) (MMCP-1). | |||||
|
PROC_HUMAN
|
||||||
| NC score | 0.090338 (rank : 127) | θ value | θ > 10 (rank : 128) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04070, Q15189, Q15190, Q16001 | Gene names | PROC | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein C precursor (EC 3.4.21.69) (Autoprothrombin IIA) (Anticoagulant protein C) (Blood coagulation factor XIV) [Contains: Vitamin K-dependent protein C light chain; Vitamin K-dependent protein C heavy chain; Activation peptide]. | |||||
|
K1KB9_MOUSE
|
||||||
| NC score | 0.089900 (rank : 128) | θ value | θ > 10 (rank : 85) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15949 | Gene names | Klk1b9, Egfbp3, Klk-9, Klk9 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b9 precursor (EC 3.4.21.35) (Tissue kallikrein-9) (Glandular kallikrein K9) (mGK-9) (Epidermal growth factor-binding protein type C) (EGF-BP C). | |||||
|
NRPN_MOUSE
|
||||||
| NC score | 0.089833 (rank : 129) | θ value | θ > 10 (rank : 125) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 186 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61955 | Gene names | Klk8, Nrpn, Prss19 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropsin precursor (EC 3.4.21.-) (NP) (Kallikrein-8). | |||||
|
K1B22_MOUSE
|
||||||
| NC score | 0.089796 (rank : 130) | θ value | θ > 10 (rank : 76) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 174 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15948 | Gene names | Klk1b22, Klk-22, Klk22 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b22 precursor (EC 3.4.21.35) (Glandular kallikrein K22) (Tissue kallikrein 22) (mGK-22) (Epidermal growth factor-binding protein type A) (EGF-BP A) (Nerve growth factor beta chain endopeptidase) (Beta-NGF-endopeptidase). | |||||
|
K1KB3_MOUSE
|
||||||
| NC score | 0.089720 (rank : 131) | θ value | θ > 10 (rank : 81) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00756 | Gene names | Klk1b3, Klk-3, Klk3, Ngfg | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b3 precursor (EC 3.4.21.35) (Glandular kallikrein K3) (Tissue kallikrein-3) (mGK-3) (7S nerve growth factor gamma chain) (Gamma-NGF) [Contains: Nerve growth factor gamma chain 1; Nerve growth factor gamma chain 2]. | |||||
|
ST14_HUMAN
|
||||||
| NC score | 0.089647 (rank : 132) | θ value | θ > 10 (rank : 138) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 409 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5Y6, Q9BS01, Q9H3S0, Q9HB36, Q9HCA3 | Gene names | ST14, PRSS14, SNC19, TADG15 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Matriptase) (Membrane-type serine protease 1) (MT-SP1) (Prostamin) (Serine protease TADG-15) (Tumor-associated differentially-expressed gene 15 protein). | |||||
|
GRAC_MOUSE
|
||||||
| NC score | 0.089639 (rank : 133) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P08882, Q61389 | Gene names | Gzmc, Ctla-5, Ctla5 | |||
|
Domain Architecture |

|
|||||
| Description | Granzyme C precursor (EC 3.4.21.-) (Cytotoxic cell protease 2) (CCP2) (B10). | |||||
|
K1B24_MOUSE
|
||||||
| NC score | 0.089581 (rank : 134) | θ value | θ > 10 (rank : 77) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61754, Q61755, Q9JM69 | Gene names | Klk1b24, Klk-24, Klk24 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b24 precursor (EC 3.4.21.35) (Glandular kallikrein K24) (Tissue kallikrein 24) (mGK-24). | |||||
|
K1B11_MOUSE
|
||||||
| NC score | 0.089581 (rank : 135) | θ value | θ > 10 (rank : 73) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15946 | Gene names | Klk1b11, Klk-11, Klk11 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b11 precursor (EC 3.4.21.35) (Tissue kallikrein-11) (Glandular kallikrein K11) (mGK-11). | |||||
|
KLK12_HUMAN
|
||||||
| NC score | 0.089345 (rank : 136) | θ value | θ > 10 (rank : 89) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKR0, Q9UKR1 | Gene names | KLK12, KLKL5 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-12 precursor (EC 3.4.21.-) (Kallikrein-like protein 5) (KLK-L5). | |||||
|
CFAI_MOUSE
|
||||||
| NC score | 0.089131 (rank : 137) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 290 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61129, Q9WU07 | Gene names | Cfi, If | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor I precursor (EC 3.4.21.45) (C3B/C4B inactivator) [Contains: Complement factor I heavy chain; Complement factor I light chain]. | |||||
|
K1B21_MOUSE
|
||||||
| NC score | 0.089049 (rank : 138) | θ value | θ > 10 (rank : 75) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q61759, Q61760, Q9JM70 | Gene names | Klk1b21, Klk-21, Klk21 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b21 precursor (EC 3.4.21.35) (Tissue kallikrein 21) (Glandular kallikrein K21) (mGK-21). | |||||
|
KLK9_HUMAN
|
||||||
| NC score | 0.088801 (rank : 139) | θ value | θ > 10 (rank : 102) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UKQ9 | Gene names | KLK9 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-9 precursor (EC 3.4.21.-) (Kallikrein-like protein 3) (KLK- L3). | |||||
|
KLK5_HUMAN
|
||||||
| NC score | 0.088611 (rank : 140) | θ value | θ > 10 (rank : 98) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y337, Q9HBG8 | Gene names | KLK5, SCTE | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-5 precursor (EC 3.4.21.-) (Stratum corneum tryptic enzyme) (Kallikrein-like protein 2) (KLK-L2). | |||||
|
K1B27_MOUSE
|
||||||
| NC score | 0.088406 (rank : 141) | θ value | θ > 10 (rank : 79) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JM71 | Gene names | Klk1b27, Klk-27, Klk27 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b27 precursor (EC 3.4.21.-) (Tissue kallikrein 27) (Glandular kallikrein K27) (mGK-27) (mKlk27). | |||||
|
ST14_MOUSE
|
||||||
| NC score | 0.088362 (rank : 142) | θ value | θ > 10 (rank : 139) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 419 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P56677 | Gene names | St14, Prss14 | |||
|
Domain Architecture |

|
|||||
| Description | Suppressor of tumorigenicity protein 14 (EC 3.4.21.-) (Serine protease 14) (Epithin). | |||||
|
FA10_HUMAN
|
||||||
| NC score | 0.087999 (rank : 143) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00742, Q14340 | Gene names | F10 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) (Stuart- Prower factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
K1KB8_MOUSE
|
||||||
| NC score | 0.087904 (rank : 144) | θ value | θ > 10 (rank : 84) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P07628 | Gene names | Klk1b8, Klk-8, Klk8 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b8 precursor (EC 3.4.21.35) (Tissue kallikrein-8) (Glandular kallikrein K8) (mGK-8). | |||||
|
PROC_MOUSE
|
||||||
| NC score | 0.087036 (rank : 145) | θ value | θ > 10 (rank : 129) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 426 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P33587, O35498, Q91WN8, Q99PC6 | Gene names | Proc | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein C precursor (EC 3.4.21.69) (Autoprothrombin IIA) (Anticoagulant protein C) (Blood coagulation factor XIV) [Contains: Vitamin K-dependent protein C light chain; Vitamin K-dependent protein C heavy chain; Activation peptide]. | |||||
|
MCPT8_MOUSE
|
||||||
| NC score | 0.086948 (rank : 146) | θ value | θ > 10 (rank : 119) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P43430, Q9R1F1 | Gene names | Mcpt8 | |||
|
Domain Architecture |

|
|||||
| Description | Mast cell protease 8 precursor (EC 3.4.21.-) (MMCP-8). | |||||
|
KLK10_HUMAN
|
||||||
| NC score | 0.086787 (rank : 147) | θ value | θ > 10 (rank : 86) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O43240, Q99920, Q9GZW9 | Gene names | KLK10, NES1, PRRSL1 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-10 precursor (EC 3.4.21.-) (Protease serine-like 1) (Normal epithelial cell-specific 1). | |||||
|
CS1A_MOUSE
|
||||||
| NC score | 0.086709 (rank : 148) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 360 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG14, Q8BJC4, Q8CH28, Q8VBY4 | Gene names | C1sa, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-A subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-A subcomponent heavy chain; Complement C1s-A subcomponent light chain]. | |||||
|
K1KB4_MOUSE
|
||||||
| NC score | 0.086626 (rank : 149) | θ value | θ > 10 (rank : 82) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00757 | Gene names | Klk1b4, Klk-4, Klk4, Ngfa | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase-like b4 precursor (7S nerve growth factor alpha chain) (Alpha-NGF). | |||||
|
KLK1_MOUSE
|
||||||
| NC score | 0.086427 (rank : 150) | θ value | θ > 10 (rank : 94) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15947, Q61855 | Gene names | Klk1, Klk-6, Klk6 | |||
|
Domain Architecture |

|
|||||
| Description | Glandular kallikrein K1 precursor (EC 3.4.21.35) (Tissue kallikrein-6) (mGK-6) (Renal kallikrein) (KAL-B). | |||||
|
EGFB2_MOUSE
|
||||||
| NC score | 0.086423 (rank : 151) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P36368, P00754 | Gene names | Egfbp2, Egfbp-2, Klk-13, Klk13 | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor-binding protein type B precursor (EC 3.4.21.35) (EGF-BP B) (Glandular kallikrein K13) (Tissue kallikrein 13) (mGK-13) (Prorenin-converting enzyme) (PRECE). | |||||
|
NRPN_HUMAN
|
||||||
| NC score | 0.086349 (rank : 152) | θ value | θ > 10 (rank : 124) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 182 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O60259, Q9HCB3, Q9UIL9, Q9UQ47 | Gene names | KLK8, NRPN, PRSS19, TADG14 | |||
|
Domain Architecture |

|
|||||
| Description | Neuropsin precursor (EC 3.4.21.-) (NP) (Kallikrein-8) (Ovasin) (Serine protease TADG-14) (Tumor-associated differentially expressed gene 14 protein). | |||||
|
K1KB1_MOUSE
|
||||||
| NC score | 0.086234 (rank : 153) | θ value | θ > 10 (rank : 80) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 180 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00755 | Gene names | Klk1b1, Klk-1, Klk1 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b1 precursor (EC 3.4.21.35) (Glandular kallikrein K1) (Tissue kallikrein-1) (mGK-1). | |||||
|
K1KB5_MOUSE
|
||||||
| NC score | 0.086184 (rank : 154) | θ value | θ > 10 (rank : 83) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P15945, Q52KM1, Q565D7 | Gene names | Klk1b5, Klk-5, Klk5 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b5 precursor (EC 3.4.21.35) (Glandular kallikrein K5) (Tissue kallikrein-5) (mGK-5). | |||||
|
K1B26_MOUSE
|
||||||
| NC score | 0.086004 (rank : 155) | θ value | θ > 10 (rank : 78) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 177 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P36369, P00753, P00754 | Gene names | Klk1b26, Klk-26, Klk26 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b26 precursor (EC 3.4.21.35) (Glandular kallikrein K26) (Tissue kallikrein 26) (mGK-26) (Prorenin-converting enzyme 2) (PRECE-2). | |||||
|
CS1B_MOUSE
|
||||||
| NC score | 0.085501 (rank : 156) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 361 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CFG8 | Gene names | C1sb, C1s | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1s-B subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s-B subcomponent heavy chain; Complement C1s-B subcomponent light chain]. | |||||
|
FA9_HUMAN
|
||||||
| NC score | 0.085314 (rank : 157) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 421 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00740 | Gene names | F9 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor IX precursor (EC 3.4.21.22) (Christmas factor) (Plasma thromboplastin component) (PTC) [Contains: Coagulation factor IXa light chain; Coagulation factor IXa heavy chain]. | |||||
|
C1S_HUMAN
|
||||||
| NC score | 0.085253 (rank : 158) | θ value | θ > 10 (rank : 15) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P09871, Q9UCU7, Q9UCU8, Q9UCU9, Q9UCV0, Q9UCV1, Q9UCV2, Q9UCV3, Q9UCV4, Q9UCV5, Q9UM14 | Gene names | C1S | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1s subcomponent precursor (EC 3.4.21.42) (C1 esterase) [Contains: Complement C1s subcomponent heavy chain; Complement C1s subcomponent light chain]. | |||||
|
FA10_MOUSE
|
||||||
| NC score | 0.085088 (rank : 159) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 435 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88947, O54740, Q99L32 | Gene names | F10 | |||
|
Domain Architecture |

|
|||||
| Description | Coagulation factor X precursor (EC 3.4.21.6) (Stuart factor) [Contains: Factor X light chain; Factor X heavy chain; Activated factor Xa heavy chain]. | |||||
|
CORIN_MOUSE
|
||||||
| NC score | 0.085051 (rank : 160) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 451 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Z319 | Gene names | Corin, Crn, Lrp4 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Low density lipoprotein receptor-related protein 4). | |||||
|
CORIN_HUMAN
|
||||||
| NC score | 0.084970 (rank : 161) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 453 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5Q5, Q9UHY2 | Gene names | CORIN, CRN, TMPRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Atrial natriuteric peptide-converting enzyme (EC 3.4.21.-) (pro-ANP- converting enzyme) (Corin) (Heart-specific serine proteinase ATC2) (Transmembrane protease, serine 10). | |||||
|
MASP1_MOUSE
|
||||||
| NC score | 0.084892 (rank : 162) | θ value | θ > 10 (rank : 110) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P98064 | Gene names | Masp1, Crarf | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) [Contains: Complement- activating component of Ra-reactive factor heavy chain; Complement- activating component of Ra-reactive factor light chain]. | |||||
|
KLK7_MOUSE
|
||||||
| NC score | 0.084617 (rank : 163) | θ value | θ > 10 (rank : 101) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91VE3, Q9R048 | Gene names | Klk7, Prss6, Scce | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kallikrein-7 precursor (EC 3.4.21.-) (Stratum corneum chymotryptic enzyme) (Thymopsin). | |||||
|
KLK14_HUMAN
|
||||||
| NC score | 0.084302 (rank : 164) | θ value | θ > 10 (rank : 91) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 179 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9P0G3 | Gene names | KLK14, KLKL6 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-14 precursor (EC 3.4.21.-) (Kallikrein-like protein 6) (KLK-L6). | |||||
|
MASP1_HUMAN
|
||||||
| NC score | 0.083587 (rank : 165) | θ value | θ > 10 (rank : 109) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 369 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P48740, O95570, Q9UF09 | Gene names | MASP1, CRARF, CRARF1, PRSS5 | |||
|
Domain Architecture |

|
|||||
| Description | Complement-activating component of Ra-reactive factor precursor (EC 3.4.21.-) (Ra-reactive factor serine protease p100) (RaRF) (Mannan-binding lectin serine protease 1) (Mannose-binding protein- associated serine protease) (MASP-1) [Contains: Complement-activating component of Ra-reactive factor heavy chain; Complement-activating component of Ra-reactive factor light chain]. | |||||
|
KLK4_HUMAN
|
||||||
| NC score | 0.083248 (rank : 166) | θ value | θ > 10 (rank : 97) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y5K2, Q4VB16, Q9GZL6, Q9UBJ6 | Gene names | KLK4, EMSP1, PRSS17, PSTS | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-4 precursor (EC 3.4.21.-) (Prostase) (Kallikrein-like protein 1) (KLK-L1) (Enamel matrix serine proteinase 1). | |||||
|
MASP2_MOUSE
|
||||||
| NC score | 0.083175 (rank : 167) | θ value | θ > 10 (rank : 112) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 382 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91WP0, Q9QXA4, Q9QXD2, Q9QXD5, Q9Z338 | Gene names | Masp2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
KLK7_HUMAN
|
||||||
| NC score | 0.082886 (rank : 168) | θ value | θ > 10 (rank : 100) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 175 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P49862, Q8N5N9, Q8NFV7 | Gene names | KLK7, PRSS6, SCCE | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein-7 precursor (EC 3.4.21.-) (hK7) (Stratum corneum chymotryptic enzyme) (hSCCE). | |||||
|
MASP2_HUMAN
|
||||||
| NC score | 0.082035 (rank : 169) | θ value | θ > 10 (rank : 111) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O00187, O75754, Q5TEQ5, Q5TER0, Q96QG4, Q9BZH0, Q9H498, Q9H499, Q9UBP3, Q9ULC7, Q9UMV3, Q9Y270 | Gene names | MASP2 | |||
|
Domain Architecture |

|
|||||
| Description | Mannan-binding lectin serine protease 2 precursor (EC 3.4.21.104) (Mannose-binding protein-associated serine protease 2) (MASP-2) (MBL- associated serine protease 2) [Contains: Mannan-binding lectin serine protease 2 A chain; Mannan-binding lectin serine protease 2 B chain]. | |||||
|
K1B16_MOUSE
|
||||||
| NC score | 0.080071 (rank : 170) | θ value | θ > 10 (rank : 74) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 178 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P04071 | Gene names | Klk1b16, Klk-16, Klk16 | |||
|
Domain Architecture |

|
|||||
| Description | Kallikrein 1-related peptidase b16 precursor (EC 3.4.21.54) (Gamma- renin, submandibular gland) (mGK-16). | |||||
|
C1RA_MOUSE
|
||||||
| NC score | 0.079045 (rank : 171) | θ value | θ > 10 (rank : 12) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 379 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CG16, Q99KI6, Q9ET60 | Gene names | C1ra, C1r | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r-A subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-A subcomponent) [Contains: Complement C1r-A subcomponent heavy chain; Complement C1r-A subcomponent light chain]. | |||||
|
C1RB_MOUSE
|
||||||
| NC score | 0.078113 (rank : 172) | θ value | θ > 10 (rank : 13) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8CFG9 | Gene names | C1rb, C1r | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Complement C1r-B subcomponent precursor (EC 3.4.21.41) (Complement component 1, r-B subcomponent) [Contains: Complement C1r-B subcomponent heavy chain; Complement C1r-B subcomponent light chain]. | |||||
|
C1R_HUMAN
|
||||||
| NC score | 0.077524 (rank : 173) | θ value | θ > 10 (rank : 14) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 364 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P00736, Q8J012 | Gene names | C1R | |||
|
Domain Architecture |

|
|||||
| Description | Complement C1r subcomponent precursor (EC 3.4.21.41) (Complement component 1, r subcomponent) [Contains: Complement C1r subcomponent heavy chain; Complement C1r subcomponent light chain]. | |||||
|
CFAB_HUMAN
|
||||||
| NC score | 0.071290 (rank : 174) | θ value | θ > 10 (rank : 19) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 251 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P00751, O15006, Q29944, Q96HX6, Q9BTF5, Q9BX92 | Gene names | CFB, BF | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) (Properdin factor B) (Glycine-rich beta glycoprotein) (GBG) (PBF2) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
CFAB_MOUSE
|
||||||
| NC score | 0.070602 (rank : 175) | θ value | θ > 10 (rank : 20) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 262 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P04186 | Gene names | Cfb, Bf, H2-Bf | |||
|
Domain Architecture |

|
|||||
| Description | Complement factor B precursor (EC 3.4.21.47) (C3/C5 convertase) [Contains: Complement factor B Ba fragment; Complement factor B Bb fragment]. | |||||
|
CO2_MOUSE
|
||||||
| NC score | 0.060499 (rank : 176) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 250 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P21180 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
CO2_HUMAN
|
||||||
| NC score | 0.056916 (rank : 177) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 258 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06681, O19694, Q13904 | Gene names | C2 | |||
|
Domain Architecture |

|
|||||
| Description | Complement C2 precursor (EC 3.4.21.43) (C3/C5 convertase) [Contains: Complement C2b fragment; Complement C2a fragment]. | |||||
|
PROZ_HUMAN
|
||||||
| NC score | 0.055069 (rank : 178) | θ value | θ > 10 (rank : 130) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P22891, Q15213 | Gene names | PROZ | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
PROZ_MOUSE
|
||||||
| NC score | 0.053031 (rank : 179) | θ value | θ > 10 (rank : 131) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 410 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CQW3 | Gene names | Proz | |||
|
Domain Architecture |

|
|||||
| Description | Vitamin K-dependent protein Z precursor. | |||||
|
BAP29_HUMAN
|
||||||
| NC score | 0.038022 (rank : 180) | θ value | 3.0926 (rank : 5) | |||
| Query Neighborhood Hits | 6 | Target Neighborhood Hits | 362 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UHQ4, O95003 | Gene names | BCAP29, BAP29 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell receptor-associated protein 29 (BCR-associated protein Bap29). | |||||