Please be patient as the page loads
|
PLCG_MOUSE
|
||||||
| SwissProt Accessions | Q6NVG1, Q66JP7 | Gene names | Agpat7, Aytl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase eta (EC 2.3.1.51) (1- AGP acyltransferase 7) (1-AGPAT 7) (Lysophosphatidic acid acyltransferase-eta) (LPAAT-eta) (Acyltransferase-like 3). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PLCG_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.998858 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q643R3, O43412, Q7Z4P4, Q8IUL7, Q8TB38 | Gene names | AGPAT7, AYTL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase eta (EC 2.3.1.51) (1- AGP acyltransferase 7) (1-AGPAT 7) (Lysophosphatidic acid acyltransferase-eta) (LPAAT-eta) (Acyltransferase-like 3). | |||||
|
PLCG_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6NVG1, Q66JP7 | Gene names | Agpat7, Aytl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase eta (EC 2.3.1.51) (1- AGP acyltransferase 7) (1-AGPAT 7) (Lysophosphatidic acid acyltransferase-eta) (LPAAT-eta) (Acyltransferase-like 3). | |||||
|
AYT1A_MOUSE
|
||||||
| θ value | 7.40825e-95 (rank : 3) | NC score | 0.826806 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BYI6 | Gene names | Aytl1a, Aytl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-A (EC 2.3.1.-). | |||||
|
AYTL1_HUMAN
|
||||||
| θ value | 5.49406e-90 (rank : 4) | NC score | 0.823383 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7L5N7, Q6MZJ6, Q9NX23 | Gene names | AYTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1 (EC 2.3.1.-). | |||||
|
PCAT1_MOUSE
|
||||||
| θ value | 9.39325e-82 (rank : 5) | NC score | 0.892957 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3TFD2, Q3TAX4, Q6NXZ6, Q8BG23, Q8BUX7, Q99JU6 | Gene names | Aytl2, Lpcat1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acylglycerophosphocholine O-acyltransferase 1 (EC 2.3.1.23) (Lung- type acyl-coa:lysophosphatidylcholine acyltransferase 1) (mLPCAT1) (Acyltransferase-like 2). | |||||
|
PCAT1_HUMAN
|
||||||
| θ value | 2.0926e-81 (rank : 6) | NC score | 0.894743 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NF37, Q1HAQ1, Q7Z4G6, Q8N3U7, Q8WUL8, Q9GZW6 | Gene names | AYTL2, LPCAT1, PFAAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acylglycerophosphocholine O-acyltransferase 1 (EC 2.3.1.23) (Lung- type acyl-coa:lysophosphatidylcholine acyltransferase 1) (Acyltransferase-like 2) (Phosphonoformate immuno-associated protein 3). | |||||
|
AYT1B_MOUSE
|
||||||
| θ value | 6.30065e-78 (rank : 7) | NC score | 0.808767 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D5U0 | Gene names | Aytl1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-B (EC 2.3.1.-). | |||||
|
PLCF_MOUSE
|
||||||
| θ value | 8.11959e-09 (rank : 8) | NC score | 0.509895 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K2C8, Q3TF78 | Gene names | Agpat6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase zeta precursor (EC 2.3.1.51) (1-AGP acyltransferase 6) (1-AGPAT 6) (Lysophosphatidic acid acyltransferase-zeta) (LPAAT-zeta). | |||||
|
PLCF_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 9) | NC score | 0.499383 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86UL3, Q86V89 | Gene names | AGPAT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase zeta precursor (EC 2.3.1.51) (1-AGP acyltransferase 6) (1-AGPAT 6) (Lysophosphatidic acid acyltransferase-zeta) (LPAAT-zeta). | |||||
|
PLCB_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 10) | NC score | 0.225500 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K3K7 | Gene names | Agpat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase beta (EC 2.3.1.51) (1- AGP acyltransferase 2) (1-AGPAT 2) (Lysophosphatidic acid acyltransferase-beta) (LPAAT-beta) (1-acylglycerol-3-phosphate O- acyltransferase 2). | |||||
|
PLCB_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 11) | NC score | 0.201060 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15120, O00516, O15106, Q9BSV7, Q9BWR7 | Gene names | AGPAT2 | |||
|
Domain Architecture |
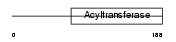
|
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase beta (EC 2.3.1.51) (1- AGP acyltransferase 2) (1-AGPAT 2) (Lysophosphatidic acid acyltransferase-beta) (LPAAT-beta) (1-acylglycerol-3-phosphate O- acyltransferase 2). | |||||
|
PLCA_HUMAN
|
||||||
| θ value | 0.163984 (rank : 12) | NC score | 0.199707 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99943, Q5BL03 | Gene names | AGPAT1, G15 | |||
|
Domain Architecture |
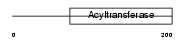
|
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase alpha (EC 2.3.1.51) (1- AGP acyltransferase 1) (1-AGPAT 1) (Lysophosphatidic acid acyltransferase-alpha) (LPAAT-alpha) (1-acylglycerol-3-phosphate O- acyltransferase 1) (Protein G15). | |||||
|
PLCA_MOUSE
|
||||||
| θ value | 0.21417 (rank : 13) | NC score | 0.163883 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35083, O35446 | Gene names | Agpat1 | |||
|
Domain Architecture |
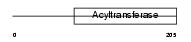
|
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase alpha (EC 2.3.1.51) (1- AGP acyltransferase 1) (1-AGPAT 1) (Lysophosphatidic acid acyltransferase-alpha) (LPAAT-alpha) (1-acylglycerol-3-phosphate O- acyltransferase 1). | |||||
|
AUP1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 14) | NC score | 0.049800 (rank : 61) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70295 | Gene names | Aup1 | |||
|
Domain Architecture |

|
|||||
| Description | Ancient ubiquitous protein 1 precursor. | |||||
|
DDR1_MOUSE
|
||||||
| θ value | 1.81305 (rank : 15) | NC score | 0.003097 (rank : 67) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 818 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03146 | Gene names | Ddr1, Cak, Eddr1, Mpk6 | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial discoidin domain-containing receptor 1 precursor (EC 2.7.10.1) (Epithelial discoidin domain receptor 1) (Tyrosine kinase DDR) (Discoidin receptor tyrosine kinase) (Tyrosine-protein kinase CAK) (Cell adhesion kinase) (Protein-tyrosine kinase MPK-6) (CD167a antigen). | |||||
|
AUP1_HUMAN
|
||||||
| θ value | 4.03905 (rank : 16) | NC score | 0.042097 (rank : 62) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y679, Q9H866, Q9UNQ6, Q9Y685 | Gene names | AUP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ancient ubiquitous protein 1 precursor. | |||||
|
DDX21_MOUSE
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.003956 (rank : 66) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
MBD5_HUMAN
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.016682 (rank : 63) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
MUCDL_MOUSE
|
||||||
| θ value | 6.88961 (rank : 19) | NC score | 0.006159 (rank : 64) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
SIG10_HUMAN
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.004167 (rank : 65) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96LC7, Q6UXI8, Q96G54, Q96LC8 | Gene names | SIGLEC10, SLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 10 precursor (Siglec-10) (Siglec- like protein 2). | |||||
|
CALL3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 21) | NC score | 0.062899 (rank : 55) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
CALL5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 22) | NC score | 0.062393 (rank : 56) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
CALM_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.056486 (rank : 59) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| θ value | θ > 10 (rank : 24) | NC score | 0.056486 (rank : 60) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CANB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.122427 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.122439 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |
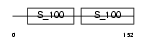
|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.132926 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
CANB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.130114 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |
|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
CSEN_HUMAN
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.077252 (rank : 39) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2W7, Q53TJ5, Q96T40, Q9UJ84, Q9UJ85 | Gene names | KCNIP3, CSEN, DREAM, KCHIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (KChIP3) (A-type potassium channel modulatory protein 3). | |||||
|
CSEN_MOUSE
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.075168 (rank : 42) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXT8, Q924L0, Q99PH9, Q99PI0, Q99PI2, Q99PI3, Q9JHZ5 | Gene names | Kcnip3, Csen, Dream, Kchip3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (A-type potassium channel modulatory protein 3) (KChIP3). | |||||
|
DUOX1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.072467 (rank : 43) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRD9, Q6ZMB3, Q6ZR09, Q9NZC1 | Gene names | DUOX1, DUOX, LNOX1, THOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 1 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH thyroid oxidase 1) (Thyroid oxidase 1) (Large NOX 1) (Long NOX 1). | |||||
|
DUOX2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.083407 (rank : 34) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRD8, Q9NR02, Q9UHF9 | Gene names | DUOX2, LNOX2, THOX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 2 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH oxidase/peroxidase DUOX2) (NADPH thyroid oxidase 2) (Thyroid oxidase 2) (NADH/NADPH thyroid oxidase p138-tox) (p138 thyroid oxidase) (Large NOX 2) (Long NOX 2). | |||||
|
EFCB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.081239 (rank : 35) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HAE3 | Gene names | EFCAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
EFCB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.065255 (rank : 52) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D3N2, Q8BSW3, Q8C982, Q8CAI0, Q9D527, Q9D5E7 | Gene names | Efcab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
GUC1A_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.105792 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43080, Q9NU14 | Gene names | GUCA1A, GCAP, GCAP1, GUCA1 | |||
|
Domain Architecture |
|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
GUC1A_MOUSE
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.104013 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43081 | Gene names | Guca1a, Gcap, Gcap1, Guca1 | |||
|
Domain Architecture |
|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
GUC1B_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.070328 (rank : 49) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UMX6, Q9NU15 | Gene names | GUCA1B, GCAP2 | |||
|
Domain Architecture |
|
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
GUC1B_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.087379 (rank : 29) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VBV8 | Gene names | Guca1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
GUC1C_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.070468 (rank : 48) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95843, O95844, Q9UNM0 | Gene names | GUCA1C, GCAP3 | |||
|
Domain Architecture |
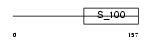
|
|||||
| Description | Guanylyl cyclase-activating protein 3 (GCAP 3) (Guanylate cyclase activator 1C). | |||||
|
HPCA_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.087921 (rank : 27) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P84074, P32076, P41211, P70510 | Gene names | HPCA, BDR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin (Calcium-binding protein BDR-2). | |||||
|
HPCA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.087921 (rank : 28) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P84075, P32076, P41211, P70510 | Gene names | Hpca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin. | |||||
|
HPCL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.089071 (rank : 26) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P37235, Q969S5 | Gene names | HPCAL1, BDR1 | |||
|
Domain Architecture |
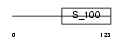
|
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Calcium-binding protein BDR-1) (HLP2). | |||||
|
HPCL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.086405 (rank : 31) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62748, P35333 | Gene names | Hpcal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Neural visinin-like protein 3) (NVL-3) (NVP-3). | |||||
|
HPCL4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.096998 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UM19, Q5TG97, Q8N611 | Gene names | HPCAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (HLP4). | |||||
|
HPCL4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.096987 (rank : 21) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BGZ1 | Gene names | Hpcal4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (Neural visinin-like protein 2) (NVP-2). | |||||
|
KCIP1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.059546 (rank : 57) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZI2, Q5U822 | Gene names | KCNIP1, KCHIP1, VABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1) (Vesicle APC-binding protein). | |||||
|
KCIP1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.059465 (rank : 58) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJ57, Q5SSA3, Q6DTJ1, Q8BGJ4, Q8C4K4, Q8CGL1, Q8K1U1, Q8K3M2 | Gene names | Kcnip1, Kchip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1). | |||||
|
KCIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.072024 (rank : 45) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NS61, Q7Z6F1, Q96K86, Q96T41, Q96T42, Q96T43, Q96T44, Q9H0N4, Q9HD10, Q9HD11, Q9NS60, Q9NY10, Q9NZI1 | Gene names | KCNIP2, KCHIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2) (Cardiac voltage-gated potassium channel modulatory subunit). | |||||
|
KCIP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 49) | NC score | 0.071252 (rank : 47) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJ69, Q6DTJ2, Q8K1T8, Q8K1T9, Q8K1U0, Q8VHN4, Q8VHN5, Q8VHN6, Q9JJ68 | Gene names | Kcnip2, Kchip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2). | |||||
|
KCIP4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 50) | NC score | 0.068444 (rank : 50) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PIL6, Q4W5G8, Q8NEU0, Q9BWT2, Q9H294, Q9H2A4 | Gene names | KCNIP4, CALP, KCHIP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
KCIP4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 51) | NC score | 0.068428 (rank : 51) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PHZ8, Q6DTJ3, Q8CAD0, Q8R4I2, Q9EQ01 | Gene names | Kcnip4, Calp, Kchip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
KIP2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 52) | NC score | 0.087302 (rank : 30) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75838 | Gene names | CIB2, KIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
KIP2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 53) | NC score | 0.076103 (rank : 40) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z309 | Gene names | Cib2, Kip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
NCALD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 54) | NC score | 0.091695 (rank : 24) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P61601, P29554, Q8IYC3, Q9H0W2 | Gene names | NCALD | |||
|
Domain Architecture |
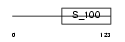
|
|||||
| Description | Neurocalcin delta. | |||||
|
NCALD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 55) | NC score | 0.091695 (rank : 25) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91X97, Q3TJS9, Q8BZN9 | Gene names | Ncald, D15Ertd412e | |||
|
Domain Architecture |
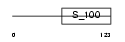
|
|||||
| Description | Neurocalcin delta. | |||||
|
NCS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 56) | NC score | 0.083570 (rank : 32) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62166, P36610, Q9UK26 | Gene names | FREQ, FLUP, NCS1 | |||
|
Domain Architecture |
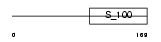
|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog) (Frequenin-like protein) (Frequenin-like ubiquitous protein). | |||||
|
NCS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 57) | NC score | 0.083570 (rank : 33) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BNY6 | Gene names | Freq, Ncs1 | |||
|
Domain Architecture |
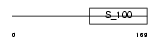
|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog). | |||||
|
NOX5_HUMAN
|
||||||
| θ value | θ > 10 (rank : 58) | NC score | 0.078736 (rank : 38) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96PH1, Q8TEQ1, Q8TER4, Q96PH2, Q96PJ8, Q96PJ9, Q9H6E0, Q9HAM8 | Gene names | NOX5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase 5 (EC 1.6.3.-). | |||||
|
RECO_HUMAN
|
||||||
| θ value | θ > 10 (rank : 59) | NC score | 0.071992 (rank : 46) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35243 | Gene names | RCVRN, RCV1 | |||
|
Domain Architecture |
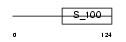
|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR). | |||||
|
RECO_MOUSE
|
||||||
| θ value | θ > 10 (rank : 60) | NC score | 0.075705 (rank : 41) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P34057 | Gene names | Rcvrn, Rcv1 | |||
|
Domain Architecture |
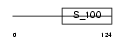
|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR) (23 kDa photoreceptor cell-specific protein). | |||||
|
TNNC1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 61) | NC score | 0.080187 (rank : 36) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
TNNC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 62) | NC score | 0.079600 (rank : 37) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |
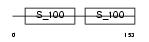
|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
TNNC2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 63) | NC score | 0.064125 (rank : 53) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |
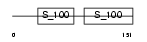
|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
TNNC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 64) | NC score | 0.062971 (rank : 54) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |
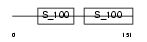
|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
UBP32_HUMAN
|
||||||
| θ value | θ > 10 (rank : 65) | NC score | 0.072202 (rank : 44) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
VISL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 66) | NC score | 0.094093 (rank : 22) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62760, P28677, P29103, P42323, Q9UM20 | Gene names | VSNL1, VISL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Hippocalcin-like protein 3) (HLP3). | |||||
|
VISL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 67) | NC score | 0.094093 (rank : 23) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62761, P28677, P29103, P42323, Q9UM20 | Gene names | Vsnl1, Visl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Neural visinin-like protein 1) (NVL-1) (NVP-1). | |||||
|
PLCG_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q6NVG1, Q66JP7 | Gene names | Agpat7, Aytl3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase eta (EC 2.3.1.51) (1- AGP acyltransferase 7) (1-AGPAT 7) (Lysophosphatidic acid acyltransferase-eta) (LPAAT-eta) (Acyltransferase-like 3). | |||||
|
PLCG_HUMAN
|
||||||
| NC score | 0.998858 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 25 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q643R3, O43412, Q7Z4P4, Q8IUL7, Q8TB38 | Gene names | AGPAT7, AYTL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase eta (EC 2.3.1.51) (1- AGP acyltransferase 7) (1-AGPAT 7) (Lysophosphatidic acid acyltransferase-eta) (LPAAT-eta) (Acyltransferase-like 3). | |||||
|
PCAT1_HUMAN
|
||||||
| NC score | 0.894743 (rank : 3) | θ value | 2.0926e-81 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 80 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8NF37, Q1HAQ1, Q7Z4G6, Q8N3U7, Q8WUL8, Q9GZW6 | Gene names | AYTL2, LPCAT1, PFAAP3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acylglycerophosphocholine O-acyltransferase 1 (EC 2.3.1.23) (Lung- type acyl-coa:lysophosphatidylcholine acyltransferase 1) (Acyltransferase-like 2) (Phosphonoformate immuno-associated protein 3). | |||||
|
PCAT1_MOUSE
|
||||||
| NC score | 0.892957 (rank : 4) | θ value | 9.39325e-82 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q3TFD2, Q3TAX4, Q6NXZ6, Q8BG23, Q8BUX7, Q99JU6 | Gene names | Aytl2, Lpcat1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acylglycerophosphocholine O-acyltransferase 1 (EC 2.3.1.23) (Lung- type acyl-coa:lysophosphatidylcholine acyltransferase 1) (mLPCAT1) (Acyltransferase-like 2). | |||||
|
AYT1A_MOUSE
|
||||||
| NC score | 0.826806 (rank : 5) | θ value | 7.40825e-95 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8BYI6 | Gene names | Aytl1a, Aytl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-A (EC 2.3.1.-). | |||||
|
AYTL1_HUMAN
|
||||||
| NC score | 0.823383 (rank : 6) | θ value | 5.49406e-90 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q7L5N7, Q6MZJ6, Q9NX23 | Gene names | AYTL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1 (EC 2.3.1.-). | |||||
|
AYT1B_MOUSE
|
||||||
| NC score | 0.808767 (rank : 7) | θ value | 6.30065e-78 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q9D5U0 | Gene names | Aytl1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Acyltransferase-like 1-B (EC 2.3.1.-). | |||||
|
PLCF_MOUSE
|
||||||
| NC score | 0.509895 (rank : 8) | θ value | 8.11959e-09 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q8K2C8, Q3TF78 | Gene names | Agpat6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase zeta precursor (EC 2.3.1.51) (1-AGP acyltransferase 6) (1-AGPAT 6) (Lysophosphatidic acid acyltransferase-zeta) (LPAAT-zeta). | |||||
|
PLCF_HUMAN
|
||||||
| NC score | 0.499383 (rank : 9) | θ value | 4.0297e-08 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q86UL3, Q86V89 | Gene names | AGPAT6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase zeta precursor (EC 2.3.1.51) (1-AGP acyltransferase 6) (1-AGPAT 6) (Lysophosphatidic acid acyltransferase-zeta) (LPAAT-zeta). | |||||
|
PLCB_MOUSE
|
||||||
| NC score | 0.225500 (rank : 10) | θ value | 0.00228821 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8K3K7 | Gene names | Agpat2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase beta (EC 2.3.1.51) (1- AGP acyltransferase 2) (1-AGPAT 2) (Lysophosphatidic acid acyltransferase-beta) (LPAAT-beta) (1-acylglycerol-3-phosphate O- acyltransferase 2). | |||||
|
PLCB_HUMAN
|
||||||
| NC score | 0.201060 (rank : 11) | θ value | 0.00869519 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | O15120, O00516, O15106, Q9BSV7, Q9BWR7 | Gene names | AGPAT2 | |||
|
Domain Architecture |

|
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase beta (EC 2.3.1.51) (1- AGP acyltransferase 2) (1-AGPAT 2) (Lysophosphatidic acid acyltransferase-beta) (LPAAT-beta) (1-acylglycerol-3-phosphate O- acyltransferase 2). | |||||
|
PLCA_HUMAN
|
||||||
| NC score | 0.199707 (rank : 12) | θ value | 0.163984 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q99943, Q5BL03 | Gene names | AGPAT1, G15 | |||
|
Domain Architecture |

|
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase alpha (EC 2.3.1.51) (1- AGP acyltransferase 1) (1-AGPAT 1) (Lysophosphatidic acid acyltransferase-alpha) (LPAAT-alpha) (1-acylglycerol-3-phosphate O- acyltransferase 1) (Protein G15). | |||||
|
PLCA_MOUSE
|
||||||
| NC score | 0.163883 (rank : 13) | θ value | 0.21417 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | O35083, O35446 | Gene names | Agpat1 | |||
|
Domain Architecture |

|
|||||
| Description | 1-acyl-sn-glycerol-3-phosphate acyltransferase alpha (EC 2.3.1.51) (1- AGP acyltransferase 1) (1-AGPAT 1) (Lysophosphatidic acid acyltransferase-alpha) (LPAAT-alpha) (1-acylglycerol-3-phosphate O- acyltransferase 1). | |||||
|
CANB2_HUMAN
|
||||||
| NC score | 0.132926 (rank : 14) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96LZ3, Q7Z4V8, Q8WYJ4 | Gene names | PPP3R2, CBLP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform) (Calcineurin B-like protein) (CBLP) (CNBII). | |||||
|
CANB2_MOUSE
|
||||||
| NC score | 0.130114 (rank : 15) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 173 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q63811 | Gene names | Ppp3r2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 2 (Protein phosphatase 2B regulatory subunit 2) (Protein phosphatase 3 regulatory subunit B beta isoform). | |||||
|
CANB1_MOUSE
|
||||||
| NC score | 0.122439 (rank : 16) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q63810 | Gene names | Ppp3r1, Cnb | |||
|
Domain Architecture |

|
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
CANB1_HUMAN
|
||||||
| NC score | 0.122427 (rank : 17) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63098, P06705, P15117, Q08044 | Gene names | PPP3R1, CNA2, CNB | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcineurin subunit B isoform 1 (Protein phosphatase 2B regulatory subunit 1) (Protein phosphatase 3 regulatory subunit B alpha isoform 1). | |||||
|
GUC1A_HUMAN
|
||||||
| NC score | 0.105792 (rank : 18) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 147 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43080, Q9NU14 | Gene names | GUCA1A, GCAP, GCAP1, GUCA1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
GUC1A_MOUSE
|
||||||
| NC score | 0.104013 (rank : 19) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P43081 | Gene names | Guca1a, Gcap, Gcap1, Guca1 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 1 (GCAP 1) (Guanylate cyclase activator 1A). | |||||
|
HPCL4_HUMAN
|
||||||
| NC score | 0.096998 (rank : 20) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9UM19, Q5TG97, Q8N611 | Gene names | HPCAL4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (HLP4). | |||||
|
HPCL4_MOUSE
|
||||||
| NC score | 0.096987 (rank : 21) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 123 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BGZ1 | Gene names | Hpcal4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 4 (Neural visinin-like protein 2) (NVP-2). | |||||
|
VISL1_HUMAN
|
||||||
| NC score | 0.094093 (rank : 22) | θ value | θ > 10 (rank : 66) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62760, P28677, P29103, P42323, Q9UM20 | Gene names | VSNL1, VISL1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Hippocalcin-like protein 3) (HLP3). | |||||
|
VISL1_MOUSE
|
||||||
| NC score | 0.094093 (rank : 23) | θ value | θ > 10 (rank : 67) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62761, P28677, P29103, P42323, Q9UM20 | Gene names | Vsnl1, Visl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Visinin-like protein 1 (VILIP) (Neural visinin-like protein 1) (NVL-1) (NVP-1). | |||||
|
NCALD_HUMAN
|
||||||
| NC score | 0.091695 (rank : 24) | θ value | θ > 10 (rank : 54) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P61601, P29554, Q8IYC3, Q9H0W2 | Gene names | NCALD | |||
|
Domain Architecture |

|
|||||
| Description | Neurocalcin delta. | |||||
|
NCALD_MOUSE
|
||||||
| NC score | 0.091695 (rank : 25) | θ value | θ > 10 (rank : 55) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q91X97, Q3TJS9, Q8BZN9 | Gene names | Ncald, D15Ertd412e | |||
|
Domain Architecture |

|
|||||
| Description | Neurocalcin delta. | |||||
|
HPCL1_HUMAN
|
||||||
| NC score | 0.089071 (rank : 26) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P37235, Q969S5 | Gene names | HPCAL1, BDR1 | |||
|
Domain Architecture |

|
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Calcium-binding protein BDR-1) (HLP2). | |||||
|
HPCA_HUMAN
|
||||||
| NC score | 0.087921 (rank : 27) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P84074, P32076, P41211, P70510 | Gene names | HPCA, BDR2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin (Calcium-binding protein BDR-2). | |||||
|
HPCA_MOUSE
|
||||||
| NC score | 0.087921 (rank : 28) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 156 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P84075, P32076, P41211, P70510 | Gene names | Hpca | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Neuron-specific calcium-binding protein hippocalcin. | |||||
|
GUC1B_MOUSE
|
||||||
| NC score | 0.087379 (rank : 29) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 116 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VBV8 | Gene names | Guca1b | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
KIP2_HUMAN
|
||||||
| NC score | 0.087302 (rank : 30) | θ value | θ > 10 (rank : 52) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 107 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O75838 | Gene names | CIB2, KIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
HPCL1_MOUSE
|
||||||
| NC score | 0.086405 (rank : 31) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 149 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62748, P35333 | Gene names | Hpcal1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Hippocalcin-like protein 1 (Visinin-like protein 3) (VILIP-3) (Neural visinin-like protein 3) (NVL-3) (NVP-3). | |||||
|
NCS1_HUMAN
|
||||||
| NC score | 0.083570 (rank : 32) | θ value | θ > 10 (rank : 56) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62166, P36610, Q9UK26 | Gene names | FREQ, FLUP, NCS1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog) (Frequenin-like protein) (Frequenin-like ubiquitous protein). | |||||
|
NCS1_MOUSE
|
||||||
| NC score | 0.083570 (rank : 33) | θ value | θ > 10 (rank : 57) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 132 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8BNY6 | Gene names | Freq, Ncs1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal calcium sensor 1 (NCS-1) (Frequenin homolog). | |||||
|
DUOX2_HUMAN
|
||||||
| NC score | 0.083407 (rank : 34) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 139 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRD8, Q9NR02, Q9UHF9 | Gene names | DUOX2, LNOX2, THOX2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 2 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH oxidase/peroxidase DUOX2) (NADPH thyroid oxidase 2) (Thyroid oxidase 2) (NADH/NADPH thyroid oxidase p138-tox) (p138 thyroid oxidase) (Large NOX 2) (Long NOX 2). | |||||
|
EFCB1_HUMAN
|
||||||
| NC score | 0.081239 (rank : 35) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 121 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9HAE3 | Gene names | EFCAB1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
TNNC1_HUMAN
|
||||||
| NC score | 0.080187 (rank : 36) | θ value | θ > 10 (rank : 61) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P63316, O14800, P02590, P04463 | Gene names | TNNC1, TNNC | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
TNNC1_MOUSE
|
||||||
| NC score | 0.079600 (rank : 37) | θ value | θ > 10 (rank : 62) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 221 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P19123 | Gene names | Tnnc1, Tncc | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, slow skeletal and cardiac muscles (TN-C). | |||||
|
NOX5_HUMAN
|
||||||
| NC score | 0.078736 (rank : 38) | θ value | θ > 10 (rank : 58) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 89 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96PH1, Q8TEQ1, Q8TER4, Q96PH2, Q96PJ8, Q96PJ9, Q9H6E0, Q9HAM8 | Gene names | NOX5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | NADPH oxidase 5 (EC 1.6.3.-). | |||||
|
CSEN_HUMAN
|
||||||
| NC score | 0.077252 (rank : 39) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 128 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Y2W7, Q53TJ5, Q96T40, Q9UJ84, Q9UJ85 | Gene names | KCNIP3, CSEN, DREAM, KCHIP3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (KChIP3) (A-type potassium channel modulatory protein 3). | |||||
|
KIP2_MOUSE
|
||||||
| NC score | 0.076103 (rank : 40) | θ value | θ > 10 (rank : 53) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 110 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9Z309 | Gene names | Cib2, Kip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Calcium and integrin-binding protein 2 (Kinase-interacting protein 2) (KIP 2). | |||||
|
RECO_MOUSE
|
||||||
| NC score | 0.075705 (rank : 41) | θ value | θ > 10 (rank : 60) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P34057 | Gene names | Rcvrn, Rcv1 | |||
|
Domain Architecture |

|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR) (23 kDa photoreceptor cell-specific protein). | |||||
|
CSEN_MOUSE
|
||||||
| NC score | 0.075168 (rank : 42) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 117 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9QXT8, Q924L0, Q99PH9, Q99PI0, Q99PI2, Q99PI3, Q9JHZ5 | Gene names | Kcnip3, Csen, Dream, Kchip3 | |||
|
Domain Architecture |

|
|||||
| Description | Calsenilin (DRE-antagonist modulator) (DREAM) (Kv channel-interacting protein 3) (A-type potassium channel modulatory protein 3) (KChIP3). | |||||
|
DUOX1_HUMAN
|
||||||
| NC score | 0.072467 (rank : 43) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NRD9, Q6ZMB3, Q6ZR09, Q9NZC1 | Gene names | DUOX1, DUOX, LNOX1, THOX1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Dual oxidase 1 precursor (EC 1.6.3.1) (EC 1.11.1.-) (NADPH thyroid oxidase 1) (Thyroid oxidase 1) (Large NOX 1) (Long NOX 1). | |||||
|
UBP32_HUMAN
|
||||||
| NC score | 0.072202 (rank : 44) | θ value | θ > 10 (rank : 65) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 170 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8NFA0, Q9BX85, Q9Y591 | Gene names | USP32, USP10 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 32 (EC 3.1.2.15) (Ubiquitin thioesterase 32) (Ubiquitin-specific-processing protease 32) (Deubiquitinating enzyme 32) (NY-REN-60 antigen). | |||||
|
KCIP2_HUMAN
|
||||||
| NC score | 0.072024 (rank : 45) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 125 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NS61, Q7Z6F1, Q96K86, Q96T41, Q96T42, Q96T43, Q96T44, Q9H0N4, Q9HD10, Q9HD11, Q9NS60, Q9NY10, Q9NZI1 | Gene names | KCNIP2, KCHIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2) (Cardiac voltage-gated potassium channel modulatory subunit). | |||||
|
RECO_HUMAN
|
||||||
| NC score | 0.071992 (rank : 46) | θ value | θ > 10 (rank : 59) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 101 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P35243 | Gene names | RCVRN, RCV1 | |||
|
Domain Architecture |

|
|||||
| Description | Recoverin (Cancer-associated retinopathy protein) (Protein CAR). | |||||
|
KCIP2_MOUSE
|
||||||
| NC score | 0.071252 (rank : 47) | θ value | θ > 10 (rank : 49) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJ69, Q6DTJ2, Q8K1T8, Q8K1T9, Q8K1U0, Q8VHN4, Q8VHN5, Q8VHN6, Q9JJ68 | Gene names | Kcnip2, Kchip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 2 (KChIP2) (A-type potassium channel modulatory protein 2) (Potassium channel-interacting protein 2). | |||||
|
GUC1C_HUMAN
|
||||||
| NC score | 0.070468 (rank : 48) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 148 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95843, O95844, Q9UNM0 | Gene names | GUCA1C, GCAP3 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 3 (GCAP 3) (Guanylate cyclase activator 1C). | |||||
|
GUC1B_HUMAN
|
||||||
| NC score | 0.070328 (rank : 49) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9UMX6, Q9NU15 | Gene names | GUCA1B, GCAP2 | |||
|
Domain Architecture |

|
|||||
| Description | Guanylyl cyclase-activating protein 2 (GCAP 2) (Guanylate cyclase activator 1B). | |||||
|
KCIP4_HUMAN
|
||||||
| NC score | 0.068444 (rank : 50) | θ value | θ > 10 (rank : 50) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PIL6, Q4W5G8, Q8NEU0, Q9BWT2, Q9H294, Q9H2A4 | Gene names | KCNIP4, CALP, KCHIP4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
KCIP4_MOUSE
|
||||||
| NC score | 0.068428 (rank : 51) | θ value | θ > 10 (rank : 51) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 136 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q6PHZ8, Q6DTJ3, Q8CAD0, Q8R4I2, Q9EQ01 | Gene names | Kcnip4, Calp, Kchip4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 4 (KChIP4) (A-type potassium channel modulatory protein 4) (Potassium channel-interacting protein 4) (Calsenilin-like protein). | |||||
|
EFCB1_MOUSE
|
||||||
| NC score | 0.065255 (rank : 52) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 126 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9D3N2, Q8BSW3, Q8C982, Q8CAI0, Q9D527, Q9D5E7 | Gene names | Efcab1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | EF-hand calcium-binding domain-containing protein 1. | |||||
|
TNNC2_HUMAN
|
||||||
| NC score | 0.064125 (rank : 53) | θ value | θ > 10 (rank : 63) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P02585 | Gene names | TNNC2 | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle. | |||||
|
TNNC2_MOUSE
|
||||||
| NC score | 0.062971 (rank : 54) | θ value | θ > 10 (rank : 64) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20801 | Gene names | Tnnc2, Tncs | |||
|
Domain Architecture |

|
|||||
| Description | Troponin C, skeletal muscle (STNC). | |||||
|
CALL3_HUMAN
|
||||||
| NC score | 0.062899 (rank : 55) | θ value | θ > 10 (rank : 21) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 225 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P27482 | Gene names | CALML3 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 3 (Calmodulin-related protein NB-1) (CaM-like protein) (CLP). | |||||
|
CALL5_HUMAN
|
||||||
| NC score | 0.062393 (rank : 56) | θ value | θ > 10 (rank : 22) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZT1, Q8IXU8 | Gene names | CALML5, CLSP | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin-like protein 5 (Calmodulin-like skin protein). | |||||
|
KCIP1_HUMAN
|
||||||
| NC score | 0.059546 (rank : 57) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9NZI2, Q5U822 | Gene names | KCNIP1, KCHIP1, VABP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1) (Vesicle APC-binding protein). | |||||
|
KCIP1_MOUSE
|
||||||
| NC score | 0.059465 (rank : 58) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9JJ57, Q5SSA3, Q6DTJ1, Q8BGJ4, Q8C4K4, Q8CGL1, Q8K1U1, Q8K3M2 | Gene names | Kcnip1, Kchip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Kv channel-interacting protein 1 (KChIP1) (A-type potassium channel modulatory protein 1) (Potassium channel-interacting protein 1). | |||||
|
CALM_HUMAN
|
||||||
| NC score | 0.056486 (rank : 59) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62158, P02593, P70667, P99014, Q13942, Q53S29, Q61379, Q61380, Q96HK3 | Gene names | CALM1, CALM, CAM, CAM1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
CALM_MOUSE
|
||||||
| NC score | 0.056486 (rank : 60) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 232 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P62204, P02593, P70667, P99014, Q61379, Q61380 | Gene names | Calm1, Calm, Cam, Cam1 | |||
|
Domain Architecture |

|
|||||
| Description | Calmodulin (CaM). | |||||
|
AUP1_MOUSE
|
||||||
| NC score | 0.049800 (rank : 61) | θ value | 1.81305 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P70295 | Gene names | Aup1 | |||
|
Domain Architecture |

|
|||||
| Description | Ancient ubiquitous protein 1 precursor. | |||||
|
AUP1_HUMAN
|
||||||
| NC score | 0.042097 (rank : 62) | θ value | 4.03905 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9Y679, Q9H866, Q9UNQ6, Q9Y685 | Gene names | AUP1 | |||
|
Domain Architecture |

|
|||||
| Description | Ancient ubiquitous protein 1 precursor. | |||||
|
MBD5_HUMAN
|
||||||
| NC score | 0.016682 (rank : 63) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 176 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9P267, Q9NUV6 | Gene names | MBD5, KIAA1461 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Methyl-CpG-binding domain protein 5 (Methyl-CpG-binding protein MBD5). | |||||
|
MUCDL_MOUSE
|
||||||
| NC score | 0.006159 (rank : 64) | θ value | 6.88961 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8VHF2, Q8CEJ3, Q9D8I9 | Gene names | Mucdhl | |||
|
Domain Architecture |

|
|||||
| Description | Mucin and cadherin-like protein precursor (Mu-protocadherin). | |||||
|
SIG10_HUMAN
|
||||||
| NC score | 0.004167 (rank : 65) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 363 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96LC7, Q6UXI8, Q96G54, Q96LC8 | Gene names | SIGLEC10, SLG2 | |||
|
Domain Architecture |

|
|||||
| Description | Sialic acid-binding Ig-like lectin 10 precursor (Siglec-10) (Siglec- like protein 2). | |||||
|
DDX21_MOUSE
|
||||||
| NC score | 0.003956 (rank : 66) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 350 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9JIK5, Q9WV45 | Gene names | Ddx21 | |||
|
Domain Architecture |

|
|||||
| Description | Nucleolar RNA helicase 2 (EC 3.6.1.-) (Nucleolar RNA helicase II) (Nucleolar RNA helicase Gu) (RH II/Gu) (Gu-alpha) (DEAD box protein 21). | |||||
|
DDR1_MOUSE
|
||||||
| NC score | 0.003097 (rank : 67) | θ value | 1.81305 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 818 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q03146 | Gene names | Ddr1, Cak, Eddr1, Mpk6 | |||
|
Domain Architecture |

|
|||||
| Description | Epithelial discoidin domain-containing receptor 1 precursor (EC 2.7.10.1) (Epithelial discoidin domain receptor 1) (Tyrosine kinase DDR) (Discoidin receptor tyrosine kinase) (Tyrosine-protein kinase CAK) (Cell adhesion kinase) (Protein-tyrosine kinase MPK-6) (CD167a antigen). | |||||