Please be patient as the page loads
|
PGRP2_MOUSE
|
||||||
| SwissProt Accessions | Q8VCS0, Q8K4I8, Q9QXZ1, Q9QXZ2 | Gene names | Pglyrp2, Pglyrpl, Pgrpl | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylmuramoyl-L-alanine amidase precursor (EC 3.5.1.28) (Peptidoglycan recognition protein long) (PGRP-L) (Peptidoglycan recognition protein 2) (TagL). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
PGRP2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.990723 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96PD5, Q96N74, Q9UC60 | Gene names | PGLYRP2, PGLYRPL, PGRPL | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylmuramoyl-L-alanine amidase precursor (EC 3.5.1.28) (Peptidoglycan recognition protein long) (PGRP-L) (Peptidoglycan recognition protein 2). | |||||
|
PGRP2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VCS0, Q8K4I8, Q9QXZ1, Q9QXZ2 | Gene names | Pglyrp2, Pglyrpl, Pgrpl | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylmuramoyl-L-alanine amidase precursor (EC 3.5.1.28) (Peptidoglycan recognition protein long) (PGRP-L) (Peptidoglycan recognition protein 2) (TagL). | |||||
|
PGRP3_HUMAN
|
||||||
| θ value | 4.72714e-33 (rank : 3) | NC score | 0.821750 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96LB9 | Gene names | PGLYRP3, PGRPIA | |||
|
Domain Architecture |
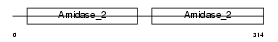
|
|||||
| Description | Peptidoglycan recognition protein I-alpha precursor (Peptidoglycan recognition protein intermediate alpha) (PGRP-I-alpha) (PGLYRPIalpha) (Peptidoglycan recognition protein 3). | |||||
|
PGRP4_HUMAN
|
||||||
| θ value | 5.77852e-31 (rank : 4) | NC score | 0.803882 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96LB8, Q9HD75 | Gene names | PGLYRP4, PGRPIB | |||
|
Domain Architecture |
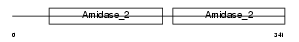
|
|||||
| Description | Peptidoglycan recognition protein I-beta precursor (Peptidoglycan recognition protein intermediate beta) (PGRP-I-beta) (PGLYRPIbeta) (Peptidoglycan recognition protein 4). | |||||
|
PGRP_HUMAN
|
||||||
| θ value | 1.85889e-29 (rank : 5) | NC score | 0.847917 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75594 | Gene names | PGLYRP1, PGLYRP, PGRP | |||
|
Domain Architecture |

|
|||||
| Description | Peptidoglycan recognition protein precursor (PGRP-S). | |||||
|
PGRP_MOUSE
|
||||||
| θ value | 3.62785e-25 (rank : 6) | NC score | 0.835570 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88593, Q62185 | Gene names | Pglyrp1, Pglyrp, Pgrp, Pgrps, Tag7 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidoglycan recognition protein precursor (Peptidoglycan recognition protein short) (PGRP-S) (Cytokine tag7). | |||||
|
PARM1_MOUSE
|
||||||
| θ value | 4.03905 (rank : 7) | NC score | 0.024892 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
MAZ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 8) | NC score | 0.000711 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56270, Q15703, Q99443 | Gene names | MAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myc-associated zinc finger protein (MAZI) (Purine-binding transcription factor) (Pur-1) (ZF87) (ZIF87). | |||||
|
MPDZ_HUMAN
|
||||||
| θ value | 5.27518 (rank : 9) | NC score | 0.007719 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
PPRB_HUMAN
|
||||||
| θ value | 6.88961 (rank : 10) | NC score | 0.013131 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
RP1L1_HUMAN
|
||||||
| θ value | 8.99809 (rank : 11) | NC score | 0.005577 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
ZDH11_HUMAN
|
||||||
| θ value | 8.99809 (rank : 12) | NC score | 0.009290 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8X9 | Gene names | ZDHHC11, ZNF399 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC11 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 11) (DHHC-11) (Zinc finger protein 399). | |||||
|
PGRP2_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8VCS0, Q8K4I8, Q9QXZ1, Q9QXZ2 | Gene names | Pglyrp2, Pglyrpl, Pgrpl | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylmuramoyl-L-alanine amidase precursor (EC 3.5.1.28) (Peptidoglycan recognition protein long) (PGRP-L) (Peptidoglycan recognition protein 2) (TagL). | |||||
|
PGRP2_HUMAN
|
||||||
| NC score | 0.990723 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96PD5, Q96N74, Q9UC60 | Gene names | PGLYRP2, PGLYRPL, PGRPL | |||
|
Domain Architecture |

|
|||||
| Description | N-acetylmuramoyl-L-alanine amidase precursor (EC 3.5.1.28) (Peptidoglycan recognition protein long) (PGRP-L) (Peptidoglycan recognition protein 2). | |||||
|
PGRP_HUMAN
|
||||||
| NC score | 0.847917 (rank : 3) | θ value | 1.85889e-29 (rank : 5) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O75594 | Gene names | PGLYRP1, PGLYRP, PGRP | |||
|
Domain Architecture |

|
|||||
| Description | Peptidoglycan recognition protein precursor (PGRP-S). | |||||
|
PGRP_MOUSE
|
||||||
| NC score | 0.835570 (rank : 4) | θ value | 3.62785e-25 (rank : 6) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O88593, Q62185 | Gene names | Pglyrp1, Pglyrp, Pgrp, Pgrps, Tag7 | |||
|
Domain Architecture |

|
|||||
| Description | Peptidoglycan recognition protein precursor (Peptidoglycan recognition protein short) (PGRP-S) (Cytokine tag7). | |||||
|
PGRP3_HUMAN
|
||||||
| NC score | 0.821750 (rank : 5) | θ value | 4.72714e-33 (rank : 3) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96LB9 | Gene names | PGLYRP3, PGRPIA | |||
|
Domain Architecture |

|
|||||
| Description | Peptidoglycan recognition protein I-alpha precursor (Peptidoglycan recognition protein intermediate alpha) (PGRP-I-alpha) (PGLYRPIalpha) (Peptidoglycan recognition protein 3). | |||||
|
PGRP4_HUMAN
|
||||||
| NC score | 0.803882 (rank : 6) | θ value | 5.77852e-31 (rank : 4) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q96LB8, Q9HD75 | Gene names | PGLYRP4, PGRPIB | |||
|
Domain Architecture |

|
|||||
| Description | Peptidoglycan recognition protein I-beta precursor (Peptidoglycan recognition protein intermediate beta) (PGRP-I-beta) (PGLYRPIbeta) (Peptidoglycan recognition protein 4). | |||||
|
PARM1_MOUSE
|
||||||
| NC score | 0.024892 (rank : 7) | θ value | 4.03905 (rank : 7) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 140 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q923D3, Q3TTV0, Q3UFU4 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein PARM-1 precursor. | |||||
|
PPRB_HUMAN
|
||||||
| NC score | 0.013131 (rank : 8) | θ value | 6.88961 (rank : 10) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 311 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q15648, O43810, O75447, Q9HD39 | Gene names | PPARBP, ARC205, DRIP205, TRAP220, TRIP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisome proliferator-activated receptor-binding protein (PBP) (PPAR-binding protein) (Thyroid hormone receptor-associated protein complex 220 kDa component) (Trap220) (Thyroid receptor-interacting protein 2) (TRIP-2) (p53 regulatory protein RB18A) (Vitamin D receptor-interacting protein complex component DRIP205) (Activator- recruited cofactor 205 kDa component) (ARC205). | |||||
|
ZDH11_HUMAN
|
||||||
| NC score | 0.009290 (rank : 9) | θ value | 8.99809 (rank : 12) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 59 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9H8X9 | Gene names | ZDHHC11, ZNF399 | |||
|
Domain Architecture |

|
|||||
| Description | Probable palmitoyltransferase ZDHHC11 (EC 2.3.1.-) (Zinc finger DHHC domain-containing protein 11) (DHHC-11) (Zinc finger protein 399). | |||||
|
MPDZ_HUMAN
|
||||||
| NC score | 0.007719 (rank : 10) | θ value | 5.27518 (rank : 9) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 261 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O75970, O43798, Q5CZ80, Q5JTX3, Q5JTX6, Q5JTX7, Q5JUC3, Q5JUC4, Q5VZ62, Q8N790 | Gene names | MPDZ, MUPP1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Multiple PDZ domain protein (Multi PDZ domain protein 1) (Multi-PDZ- domain protein 1). | |||||
|
RP1L1_HUMAN
|
||||||
| NC score | 0.005577 (rank : 11) | θ value | 8.99809 (rank : 11) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 1072 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q8IWN7, Q86SQ1, Q8IWN8, Q8IWN9, Q8IWP0, Q8IWP1, Q8IWP2 | Gene names | RP1L1 | |||
|
Domain Architecture |

|
|||||
| Description | Retinitis pigmentosa 1-like 1 protein. | |||||
|
MAZ_HUMAN
|
||||||
| NC score | 0.000711 (rank : 12) | θ value | 5.27518 (rank : 8) | |||
| Query Neighborhood Hits | 12 | Target Neighborhood Hits | 757 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P56270, Q15703, Q99443 | Gene names | MAZ | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myc-associated zinc finger protein (MAZI) (Purine-binding transcription factor) (Pur-1) (ZF87) (ZIF87). | |||||