Please be patient as the page loads
|
OXA1L_HUMAN
|
||||||
| SwissProt Accessions | Q15070 | Gene names | OXA1L | |||
|
Domain Architecture |
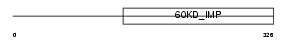
|
|||||
| Description | Inner membrane protein OXA1L, mitochondrial precursor (Oxidase assembly 1-like protein) (OXA1-like protein) (OXA1Hs) (Hsa). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
OXA1L_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15070 | Gene names | OXA1L | |||
|
Domain Architecture |

|
|||||
| Description | Inner membrane protein OXA1L, mitochondrial precursor (Oxidase assembly 1-like protein) (OXA1-like protein) (OXA1Hs) (Hsa). | |||||
|
OXA1L_MOUSE
|
||||||
| θ value | 1.41267e-170 (rank : 2) | NC score | 0.995321 (rank : 2) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGA9, Q8BK01, Q8R091, Q9D8X7 | Gene names | Oxa1l | |||
|
Domain Architecture |

|
|||||
| Description | Inner membrane protein OXA1L, mitochondrial precursor (Oxidase assembly 1-like protein) (OXA1-like protein). | |||||
|
OXAL2_HUMAN
|
||||||
| θ value | 1.2077e-20 (rank : 3) | NC score | 0.687701 (rank : 3) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8Q8 | Gene names | OXA1L2 | |||
|
Domain Architecture |
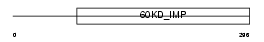
|
|||||
| Description | Probable inner membrane protein OXA1L2, mitochondrial precursor. | |||||
|
OXAL2_MOUSE
|
||||||
| θ value | 1.92812e-18 (rank : 4) | NC score | 0.682455 (rank : 4) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VC74, Q3U1D5 | Gene names | Oxa1l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable inner membrane protein OXA1L2, mitochondrial precursor. | |||||
|
TRI13_MOUSE
|
||||||
| θ value | 2.36792 (rank : 5) | NC score | 0.014540 (rank : 7) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CYB0, Q8CEV0, Q923J0, Q925P1, Q99PQ0 | Gene names | Trim13, Rfp2 | |||
|
Domain Architecture |
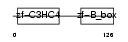
|
|||||
| Description | Tripartite motif-containing protein 13 (Ret finger protein 2) (Putative tumor suppressor RFP2). | |||||
|
GAK_MOUSE
|
||||||
| θ value | 6.88961 (rank : 6) | NC score | 0.002966 (rank : 8) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||
|
UBP10_HUMAN
|
||||||
| θ value | 8.99809 (rank : 7) | NC score | 0.015546 (rank : 5) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP10_MOUSE
|
||||||
| θ value | 8.99809 (rank : 8) | NC score | 0.015437 (rank : 6) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
OXA1L_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q15070 | Gene names | OXA1L | |||
|
Domain Architecture |

|
|||||
| Description | Inner membrane protein OXA1L, mitochondrial precursor (Oxidase assembly 1-like protein) (OXA1-like protein) (OXA1Hs) (Hsa). | |||||
|
OXA1L_MOUSE
|
||||||
| NC score | 0.995321 (rank : 2) | θ value | 1.41267e-170 (rank : 2) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BGA9, Q8BK01, Q8R091, Q9D8X7 | Gene names | Oxa1l | |||
|
Domain Architecture |

|
|||||
| Description | Inner membrane protein OXA1L, mitochondrial precursor (Oxidase assembly 1-like protein) (OXA1-like protein). | |||||
|
OXAL2_HUMAN
|
||||||
| NC score | 0.687701 (rank : 3) | θ value | 1.2077e-20 (rank : 3) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 4 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8N8Q8 | Gene names | OXA1L2 | |||
|
Domain Architecture |

|
|||||
| Description | Probable inner membrane protein OXA1L2, mitochondrial precursor. | |||||
|
OXAL2_MOUSE
|
||||||
| NC score | 0.682455 (rank : 4) | θ value | 1.92812e-18 (rank : 4) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8VC74, Q3U1D5 | Gene names | Oxa1l2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable inner membrane protein OXA1L2, mitochondrial precursor. | |||||
|
UBP10_HUMAN
|
||||||
| NC score | 0.015546 (rank : 5) | θ value | 8.99809 (rank : 7) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 153 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14694, Q9BWG7, Q9NSL7 | Gene names | USP10, KIAA0190 | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
UBP10_MOUSE
|
||||||
| NC score | 0.015437 (rank : 6) | θ value | 8.99809 (rank : 8) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P52479, Q91VY7 | Gene names | Usp10, Ode-1, Uchrp | |||
|
Domain Architecture |

|
|||||
| Description | Ubiquitin carboxyl-terminal hydrolase 10 (EC 3.1.2.15) (Ubiquitin thioesterase 10) (Ubiquitin-specific-processing protease 10) (Deubiquitinating enzyme 10). | |||||
|
TRI13_MOUSE
|
||||||
| NC score | 0.014540 (rank : 7) | θ value | 2.36792 (rank : 5) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 282 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9CYB0, Q8CEV0, Q923J0, Q925P1, Q99PQ0 | Gene names | Trim13, Rfp2 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 13 (Ret finger protein 2) (Putative tumor suppressor RFP2). | |||||
|
GAK_MOUSE
|
||||||
| NC score | 0.002966 (rank : 8) | θ value | 6.88961 (rank : 6) | |||
| Query Neighborhood Hits | 8 | Target Neighborhood Hits | 964 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q99KY4, Q6P1I8, Q6P9S5, Q8BM74, Q8K0Q4 | Gene names | Gak | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cyclin G-associated kinase (EC 2.7.11.1). | |||||