Please be patient as the page loads
|
ONCM_MOUSE
|
||||||
| SwissProt Accessions | P53347 | Gene names | Osm | |||
|
Domain Architecture |
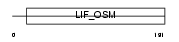
|
|||||
| Description | Oncostatin M precursor (OSM). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
ONCM_MOUSE
|
||||||
| θ value | 1.10445e-98 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53347 | Gene names | Osm | |||
|
Domain Architecture |

|
|||||
| Description | Oncostatin M precursor (OSM). | |||||
|
ONCM_HUMAN
|
||||||
| θ value | 5.95217e-44 (rank : 2) | NC score | 0.928132 (rank : 2) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13725 | Gene names | OSM | |||
|
Domain Architecture |
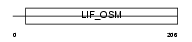
|
|||||
| Description | Oncostatin M precursor (OSM). | |||||
|
TSP1_HUMAN
|
||||||
| θ value | 2.36792 (rank : 3) | NC score | 0.014562 (rank : 5) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07996, Q15667 | Gene names | THBS1, TSP, TSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||
|
NUD12_MOUSE
|
||||||
| θ value | 4.03905 (rank : 4) | NC score | 0.022628 (rank : 4) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DCN1, Q6PFA5 | Gene names | Nudt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
CRSP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 5) | NC score | 0.026865 (rank : 3) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
ONCM_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.10445e-98 (rank : 1) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P53347 | Gene names | Osm | |||
|
Domain Architecture |

|
|||||
| Description | Oncostatin M precursor (OSM). | |||||
|
ONCM_HUMAN
|
||||||
| NC score | 0.928132 (rank : 2) | θ value | 5.95217e-44 (rank : 2) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P13725 | Gene names | OSM | |||
|
Domain Architecture |
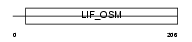
|
|||||
| Description | Oncostatin M precursor (OSM). | |||||
|
CRSP2_HUMAN
|
||||||
| NC score | 0.026865 (rank : 3) | θ value | 6.88961 (rank : 5) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 380 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O60244, Q9UNB3 | Gene names | CRSP2, ARC150, DRIP150, EXLM1, TRAP170 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CRSP complex subunit 2 (Cofactor required for Sp1 transcriptional activation subunit 2) (Transcriptional coactivator CRSP150) (Vitamin D3 receptor-interacting protein complex 150 kDa component) (DRIP150) (Thyroid hormone receptor-associated protein complex 170 kDa component) (Trap170) (Activator-recruited cofactor 150 kDa component) (ARC150). | |||||
|
NUD12_MOUSE
|
||||||
| NC score | 0.022628 (rank : 4) | θ value | 4.03905 (rank : 4) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 243 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9DCN1, Q6PFA5 | Gene names | Nudt12 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Peroxisomal NADH pyrophosphatase NUDT12 (EC 3.6.1.22) (Nucleoside diphosphate-linked moiety X motif 12) (Nudix motif 12). | |||||
|
TSP1_HUMAN
|
||||||
| NC score | 0.014562 (rank : 5) | θ value | 2.36792 (rank : 3) | |||
| Query Neighborhood Hits | 5 | Target Neighborhood Hits | 389 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P07996, Q15667 | Gene names | THBS1, TSP, TSP1 | |||
|
Domain Architecture |

|
|||||
| Description | Thrombospondin-1 precursor. | |||||