Please be patient as the page loads
|
NPTX2_HUMAN
|
||||||
| SwissProt Accessions | P47972, Q86XV7, Q96G70 | Gene names | NPTX2 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal pentraxin-2 precursor (NP2) (Neuronal pentraxin II) (NP-II). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
NPTX2_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P47972, Q86XV7, Q96G70 | Gene names | NPTX2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin-2 precursor (NP2) (Neuronal pentraxin II) (NP-II). | |||||
|
NPTX2_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.993224 (rank : 2) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O70340 | Gene names | Nptx2 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal pentraxin-2 precursor (NP2) (Neuronal pentraxin II) (NP-II). | |||||
|
NPTX1_MOUSE
|
||||||
| θ value | 1.38424e-125 (rank : 3) | NC score | 0.969341 (rank : 4) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62443 | Gene names | Nptx1 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal pentraxin-1 precursor (NP1) (Neuronal pentraxin I) (NP-I). | |||||
|
NPTX1_HUMAN
|
||||||
| θ value | 2.61056e-124 (rank : 4) | NC score | 0.970917 (rank : 3) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15818, Q5FWE6 | Gene names | NPTX1 | |||
|
Domain Architecture |
|
|||||
| Description | Neuronal pentraxin-1 precursor (NP1) (Neuronal pentraxin I) (NP-I). | |||||
|
NPTXR_MOUSE
|
||||||
| θ value | 2.9938e-112 (rank : 5) | NC score | 0.961820 (rank : 6) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99J85 | Gene names | Nptxr, Npr | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin receptor. | |||||
|
NPTXR_HUMAN
|
||||||
| θ value | 7.37397e-111 (rank : 6) | NC score | 0.967804 (rank : 5) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95502 | Gene names | NPTXR | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin receptor. | |||||
|
GP144_HUMAN
|
||||||
| θ value | 3.62785e-25 (rank : 7) | NC score | 0.413840 (rank : 13) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z7M1, Q86SL4, Q8NH12 | Gene names | GPR144, PGR24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 144 (G-protein coupled receptor PGR24). | |||||
|
PTX3_MOUSE
|
||||||
| θ value | 1.29031e-22 (rank : 8) | NC score | 0.768968 (rank : 7) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P48759, Q8VDF1 | Gene names | Ptx3, Tsg14 | |||
|
Domain Architecture |
|
|||||
| Description | Pentraxin-related protein PTX3 precursor (Pentaxin-related protein PTX3) (Tumor necrosis factor-inducible protein TSG-14). | |||||
|
PTX3_HUMAN
|
||||||
| θ value | 1.09232e-21 (rank : 9) | NC score | 0.764063 (rank : 8) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26022 | Gene names | PTX3, TSG14 | |||
|
Domain Architecture |
|
|||||
| Description | Pentraxin-related protein PTX3 precursor (Pentaxin-related protein PTX3) (Tumor necrosis factor-inducible protein TSG-14). | |||||
|
SAMP_HUMAN
|
||||||
| θ value | 9.90251e-15 (rank : 10) | NC score | 0.692641 (rank : 11) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P02743 | Gene names | APCS, PTX2 | |||
|
Domain Architecture |
|
|||||
| Description | Serum amyloid P-component precursor (SAP) (9.5S alpha-1-glycoprotein) [Contains: Serum amyloid P-component(1-203)]. | |||||
|
SAMP_MOUSE
|
||||||
| θ value | 2.20605e-14 (rank : 11) | NC score | 0.690573 (rank : 12) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12246 | Gene names | Apcs, Ptx2, Sap | |||
|
Domain Architecture |
|
|||||
| Description | Serum amyloid P-component precursor (SAP). | |||||
|
CRP_MOUSE
|
||||||
| θ value | 4.91457e-14 (rank : 12) | NC score | 0.703589 (rank : 9) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P14847 | Gene names | Crp, Ptx1 | |||
|
Domain Architecture |
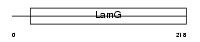
|
|||||
| Description | C-reactive protein precursor. | |||||
|
CRP_HUMAN
|
||||||
| θ value | 6.41864e-14 (rank : 13) | NC score | 0.698780 (rank : 10) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P02741, Q8WW75 | Gene names | CRP, PTX1 | |||
|
Domain Architecture |
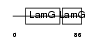
|
|||||
| Description | C-reactive protein precursor [Contains: C-reactive protein(1-205)]. | |||||
|
GP112_HUMAN
|
||||||
| θ value | 0.00020696 (rank : 14) | NC score | 0.111426 (rank : 14) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
CP250_MOUSE
|
||||||
| θ value | 0.0431538 (rank : 15) | NC score | 0.052600 (rank : 21) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
MYOC_HUMAN
|
||||||
| θ value | 0.0736092 (rank : 16) | NC score | 0.052333 (rank : 22) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
CF060_HUMAN
|
||||||
| θ value | 0.0961366 (rank : 17) | NC score | 0.053702 (rank : 20) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
CENPF_HUMAN
|
||||||
| θ value | 0.125558 (rank : 18) | NC score | 0.053822 (rank : 19) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
GCC2_MOUSE
|
||||||
| θ value | 0.125558 (rank : 19) | NC score | 0.054655 (rank : 18) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
MYH9_MOUSE
|
||||||
| θ value | 0.125558 (rank : 20) | NC score | 0.044769 (rank : 27) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_HUMAN
|
||||||
| θ value | 0.163984 (rank : 21) | NC score | 0.043279 (rank : 28) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
GCC2_HUMAN
|
||||||
| θ value | 0.21417 (rank : 22) | NC score | 0.051374 (rank : 26) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
RBP24_HUMAN
|
||||||
| θ value | 0.21417 (rank : 23) | NC score | 0.051469 (rank : 25) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
DDIT3_HUMAN
|
||||||
| θ value | 0.279714 (rank : 24) | NC score | 0.059273 (rank : 17) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35638 | Gene names | DDIT3, CHOP, GADD153 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
BPA1_MOUSE
|
||||||
| θ value | 0.365318 (rank : 25) | NC score | 0.038418 (rank : 43) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MYH11_HUMAN
|
||||||
| θ value | 0.365318 (rank : 26) | NC score | 0.041609 (rank : 33) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
TRI32_MOUSE
|
||||||
| θ value | 0.365318 (rank : 27) | NC score | 0.028662 (rank : 59) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CH72, Q8K055 | Gene names | Trim32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-). | |||||
|
GFAP_HUMAN
|
||||||
| θ value | 0.62314 (rank : 28) | NC score | 0.035818 (rank : 46) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P14136, Q53H98, Q5D055, Q6ZQS3, Q7Z5J6, Q7Z5J7, Q96KS4, Q96P18, Q9UFD0 | Gene names | GFAP | |||
|
Domain Architecture |
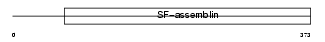
|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
BICD2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 29) | NC score | 0.042802 (rank : 29) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
FARP2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 30) | NC score | 0.011381 (rank : 80) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
MACF1_MOUSE
|
||||||
| θ value | 0.813845 (rank : 31) | NC score | 0.037645 (rank : 44) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
SAS6_MOUSE
|
||||||
| θ value | 0.813845 (rank : 32) | NC score | 0.040446 (rank : 36) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
TRIM3_HUMAN
|
||||||
| θ value | 0.813845 (rank : 33) | NC score | 0.022406 (rank : 66) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75382, Q9C038, Q9C039 | Gene names | TRIM3, BERP, RNF22, RNF97 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 3 (RING finger protein 22) (Brain- expressed RING finger protein) (RING finger protein 97). | |||||
|
ANR35_HUMAN
|
||||||
| θ value | 1.06291 (rank : 34) | NC score | 0.022500 (rank : 65) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
GP126_HUMAN
|
||||||
| θ value | 1.06291 (rank : 35) | NC score | 0.069364 (rank : 15) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86SQ4, Q6MZU7, Q8NC14, Q96JW0 | Gene names | GPR126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 126 precursor. | |||||
|
KGP2_HUMAN
|
||||||
| θ value | 1.06291 (rank : 36) | NC score | 0.005437 (rank : 83) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13237, O00125, O60916 | Gene names | PRKG2, PRKGR2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
KRHB1_HUMAN
|
||||||
| θ value | 1.06291 (rank : 37) | NC score | 0.028753 (rank : 58) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14533, Q14846, Q16274, Q8WU52, Q9BR74 | Gene names | KRTHB1, MLN137 | |||
|
Domain Architecture |
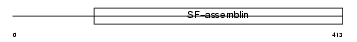
|
|||||
| Description | Keratin, type II cuticular Hb1 (Hair keratin, type II Hb1) (ghHKb1) (ghHb1) (MLN 137). | |||||
|
KRHB3_HUMAN
|
||||||
| θ value | 1.06291 (rank : 38) | NC score | 0.028550 (rank : 60) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P78385, Q9NSB3 | Gene names | KRTHB3 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cuticular Hb3 (Hair keratin, type II Hb3). | |||||
|
KRHB6_HUMAN
|
||||||
| θ value | 1.06291 (rank : 39) | NC score | 0.028873 (rank : 57) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43790, P78387 | Gene names | KRTHB6 | |||
|
Domain Architecture |
|
|||||
| Description | Keratin, type II cuticular Hb6 (Hair keratin, type II Hb6) (ghHb6). | |||||
|
CGRE1_HUMAN
|
||||||
| θ value | 1.38821 (rank : 40) | NC score | 0.019178 (rank : 71) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
EMIL2_MOUSE
|
||||||
| θ value | 1.38821 (rank : 41) | NC score | 0.037284 (rank : 45) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||
|
DOCK6_HUMAN
|
||||||
| θ value | 1.81305 (rank : 42) | NC score | 0.016882 (rank : 75) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
GFAP_MOUSE
|
||||||
| θ value | 1.81305 (rank : 43) | NC score | 0.033420 (rank : 51) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P03995, Q7TQ30, Q925K3 | Gene names | Gfap | |||
|
Domain Architecture |
|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
KINH_HUMAN
|
||||||
| θ value | 1.81305 (rank : 44) | NC score | 0.030859 (rank : 54) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
BICD2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 45) | NC score | 0.042668 (rank : 30) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
CROCC_HUMAN
|
||||||
| θ value | 2.36792 (rank : 46) | NC score | 0.051659 (rank : 24) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
KRHB6_MOUSE
|
||||||
| θ value | 2.36792 (rank : 47) | NC score | 0.026658 (rank : 62) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P97861, Q3TTV9 | Gene names | Krthb6, Krt2-10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cuticular Hb6 (Hair keratin, type II Hb6). | |||||
|
MYH8_HUMAN
|
||||||
| θ value | 2.36792 (rank : 48) | NC score | 0.033980 (rank : 50) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
SRGP2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 49) | NC score | 0.018729 (rank : 72) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
TRIM3_MOUSE
|
||||||
| θ value | 2.36792 (rank : 50) | NC score | 0.017788 (rank : 73) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R1R2, Q3UMU5 | Gene names | Trim3, Hac1, Rnf22 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 3 (RING finger protein 22) (RING finger protein HAC1). | |||||
|
CUTL1_MOUSE
|
||||||
| θ value | 3.0926 (rank : 51) | NC score | 0.031554 (rank : 52) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
MATN4_HUMAN
|
||||||
| θ value | 3.0926 (rank : 52) | NC score | 0.004250 (rank : 84) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95460, Q5QPU2, Q5QPU3, Q5QPU4, Q8N2M5, Q8N2M7, Q9H1F8, Q9H1F9 | Gene names | MATN4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor. | |||||
|
MRCKG_HUMAN
|
||||||
| θ value | 3.0926 (rank : 53) | NC score | 0.012524 (rank : 77) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
NFH_HUMAN
|
||||||
| θ value | 3.0926 (rank : 54) | NC score | 0.025717 (rank : 63) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
SPTN5_HUMAN
|
||||||
| θ value | 3.0926 (rank : 55) | NC score | 0.030368 (rank : 55) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
CCD39_HUMAN
|
||||||
| θ value | 4.03905 (rank : 56) | NC score | 0.041317 (rank : 34) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CENPE_HUMAN
|
||||||
| θ value | 4.03905 (rank : 57) | NC score | 0.034517 (rank : 48) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CNTP4_HUMAN
|
||||||
| θ value | 4.03905 (rank : 58) | NC score | 0.011588 (rank : 78) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
GOGA2_HUMAN
|
||||||
| θ value | 4.03905 (rank : 59) | NC score | 0.042445 (rank : 31) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
IFT81_MOUSE
|
||||||
| θ value | 4.03905 (rank : 60) | NC score | 0.035745 (rank : 47) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
KGP2_MOUSE
|
||||||
| θ value | 4.03905 (rank : 61) | NC score | 0.004024 (rank : 85) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61410 | Gene names | Prkg2, Prkgr2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
PCNT_HUMAN
|
||||||
| θ value | 4.03905 (rank : 62) | NC score | 0.051779 (rank : 23) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
TPD54_HUMAN
|
||||||
| θ value | 4.03905 (rank : 63) | NC score | 0.030304 (rank : 56) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43399, Q9H3Z6 | Gene names | TPD52L2 | |||
|
Domain Architecture |
|
|||||
| Description | Tumor protein D54 (hD54) (Tumor protein D52-like 2). | |||||
|
CCD46_MOUSE
|
||||||
| θ value | 5.27518 (rank : 64) | NC score | 0.040179 (rank : 37) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
FLIP1_MOUSE
|
||||||
| θ value | 5.27518 (rank : 65) | NC score | 0.042036 (rank : 32) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
FNBP2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 66) | NC score | 0.020095 (rank : 70) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
LRRF2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 67) | NC score | 0.039586 (rank : 38) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91WK0, Q8BVD1, Q9CU89 | Gene names | Lrrfip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
ROCK2_MOUSE
|
||||||
| θ value | 5.27518 (rank : 68) | NC score | 0.017728 (rank : 74) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
CASP_MOUSE
|
||||||
| θ value | 6.88961 (rank : 69) | NC score | 0.034194 (rank : 49) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
CNTP4_MOUSE
|
||||||
| θ value | 6.88961 (rank : 70) | NC score | 0.009929 (rank : 82) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99P47 | Gene names | Cntnap4, Caspr4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
FLIP1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 71) | NC score | 0.039249 (rank : 39) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
GLE1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 72) | NC score | 0.025183 (rank : 64) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q53GS7, O75458, Q53GT9, Q5VVU1, Q8NCP6, Q9UFL6 | Gene names | GLE1L, GLE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein) (hGLE1). | |||||
|
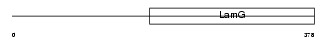
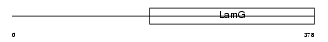
LAMB2_MOUSE
|
||||||
| θ value | 6.88961 (rank : 73) | NC score | 0.010303 (rank : 81) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
LRRF1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 74) | NC score | 0.027013 (rank : 61) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
MYH11_MOUSE
|
||||||
| θ value | 6.88961 (rank : 75) | NC score | 0.038708 (rank : 41) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
RB6I2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 76) | NC score | 0.038865 (rank : 40) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
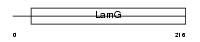
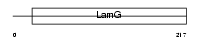
TENA_HUMAN
|
||||||
| θ value | 6.88961 (rank : 77) | NC score | 0.001870 (rank : 87) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||
|
TRI32_HUMAN
|
||||||
| θ value | 6.88961 (rank : 78) | NC score | 0.021698 (rank : 68) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13049, Q9NQP8 | Gene names | TRIM32, HT2A | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-) (Zinc finger protein HT2A) (72 kDa Tat-interacting protein). | |||||
|
APXL_HUMAN
|
||||||
| θ value | 8.99809 (rank : 79) | NC score | 0.011525 (rank : 79) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
CD72_HUMAN
|
||||||
| θ value | 8.99809 (rank : 80) | NC score | 0.020542 (rank : 69) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P21854 | Gene names | CD72 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell differentiation antigen CD72 (Lyb-2). | |||||
|
CING_MOUSE
|
||||||
| θ value | 8.99809 (rank : 81) | NC score | 0.040953 (rank : 35) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
COBA2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 82) | NC score | 0.001943 (rank : 86) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
NUCB2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 83) | NC score | 0.022388 (rank : 67) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
REST_MOUSE
|
||||||
| θ value | 8.99809 (rank : 84) | NC score | 0.038478 (rank : 42) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
RIMB1_MOUSE
|
||||||
| θ value | 8.99809 (rank : 85) | NC score | 0.031155 (rank : 53) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
SRGP2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 86) | NC score | 0.015754 (rank : 76) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
GP133_HUMAN
|
||||||
| θ value | θ > 10 (rank : 87) | NC score | 0.066952 (rank : 16) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6QNK2, Q6ZMQ1, Q7Z7M2, Q86SM4 | Gene names | GPR133, PGR25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 133 precursor (G-protein coupled receptor PGR25). | |||||
|
NPTX2_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 86 | Shared Neighborhood Hits | 86 | |
| SwissProt Accessions | P47972, Q86XV7, Q96G70 | Gene names | NPTX2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin-2 precursor (NP2) (Neuronal pentraxin II) (NP-II). | |||||
|
NPTX2_MOUSE
|
||||||
| NC score | 0.993224 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 79 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | O70340 | Gene names | Nptx2 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin-2 precursor (NP2) (Neuronal pentraxin II) (NP-II). | |||||
|
NPTX1_HUMAN
|
||||||
| NC score | 0.970917 (rank : 3) | θ value | 2.61056e-124 (rank : 4) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 57 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q15818, Q5FWE6 | Gene names | NPTX1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin-1 precursor (NP1) (Neuronal pentraxin I) (NP-I). | |||||
|
NPTX1_MOUSE
|
||||||
| NC score | 0.969341 (rank : 4) | θ value | 1.38424e-125 (rank : 3) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 67 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | Q62443 | Gene names | Nptx1 | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin-1 precursor (NP1) (Neuronal pentraxin I) (NP-I). | |||||
|
NPTXR_HUMAN
|
||||||
| NC score | 0.967804 (rank : 5) | θ value | 7.37397e-111 (rank : 6) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 60 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | O95502 | Gene names | NPTXR | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin receptor. | |||||
|
NPTXR_MOUSE
|
||||||
| NC score | 0.961820 (rank : 6) | θ value | 2.9938e-112 (rank : 5) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 87 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q99J85 | Gene names | Nptxr, Npr | |||
|
Domain Architecture |

|
|||||
| Description | Neuronal pentraxin receptor. | |||||
|
PTX3_MOUSE
|
||||||
| NC score | 0.768968 (rank : 7) | θ value | 1.29031e-22 (rank : 8) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | P48759, Q8VDF1 | Gene names | Ptx3, Tsg14 | |||
|
Domain Architecture |

|
|||||
| Description | Pentraxin-related protein PTX3 precursor (Pentaxin-related protein PTX3) (Tumor necrosis factor-inducible protein TSG-14). | |||||
|
PTX3_HUMAN
|
||||||
| NC score | 0.764063 (rank : 8) | θ value | 1.09232e-21 (rank : 9) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | P26022 | Gene names | PTX3, TSG14 | |||
|
Domain Architecture |

|
|||||
| Description | Pentraxin-related protein PTX3 precursor (Pentaxin-related protein PTX3) (Tumor necrosis factor-inducible protein TSG-14). | |||||
|
CRP_MOUSE
|
||||||
| NC score | 0.703589 (rank : 9) | θ value | 4.91457e-14 (rank : 12) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P14847 | Gene names | Crp, Ptx1 | |||
|
Domain Architecture |

|
|||||
| Description | C-reactive protein precursor. | |||||
|
CRP_HUMAN
|
||||||
| NC score | 0.698780 (rank : 10) | θ value | 6.41864e-14 (rank : 13) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P02741, Q8WW75 | Gene names | CRP, PTX1 | |||
|
Domain Architecture |

|
|||||
| Description | C-reactive protein precursor [Contains: C-reactive protein(1-205)]. | |||||
|
SAMP_HUMAN
|
||||||
| NC score | 0.692641 (rank : 11) | θ value | 9.90251e-15 (rank : 10) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P02743 | Gene names | APCS, PTX2 | |||
|
Domain Architecture |

|
|||||
| Description | Serum amyloid P-component precursor (SAP) (9.5S alpha-1-glycoprotein) [Contains: Serum amyloid P-component(1-203)]. | |||||
|
SAMP_MOUSE
|
||||||
| NC score | 0.690573 (rank : 12) | θ value | 2.20605e-14 (rank : 11) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | P12246 | Gene names | Apcs, Ptx2, Sap | |||
|
Domain Architecture |

|
|||||
| Description | Serum amyloid P-component precursor (SAP). | |||||
|
GP144_HUMAN
|
||||||
| NC score | 0.413840 (rank : 13) | θ value | 3.62785e-25 (rank : 7) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 108 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | Q7Z7M1, Q86SL4, Q8NH12 | Gene names | GPR144, PGR24 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 144 (G-protein coupled receptor PGR24). | |||||
|
GP112_HUMAN
|
||||||
| NC score | 0.111426 (rank : 14) | θ value | 0.00020696 (rank : 14) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 441 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | Q8IZF6, Q86SM6 | Gene names | GPR112 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 112. | |||||
|
GP126_HUMAN
|
||||||
| NC score | 0.069364 (rank : 15) | θ value | 1.06291 (rank : 35) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 11 | |
| SwissProt Accessions | Q86SQ4, Q6MZU7, Q8NC14, Q96JW0 | Gene names | GPR126 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 126 precursor. | |||||
|
GP133_HUMAN
|
||||||
| NC score | 0.066952 (rank : 16) | θ value | θ > 10 (rank : 87) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 115 | Shared Neighborhood Hits | 10 | |
| SwissProt Accessions | Q6QNK2, Q6ZMQ1, Q7Z7M2, Q86SM4 | Gene names | GPR133, PGR25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable G-protein coupled receptor 133 precursor (G-protein coupled receptor PGR25). | |||||
|
DDIT3_HUMAN
|
||||||
| NC score | 0.059273 (rank : 17) | θ value | 0.279714 (rank : 24) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 167 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | P35638 | Gene names | DDIT3, CHOP, GADD153 | |||
|
Domain Architecture |

|
|||||
| Description | DNA damage-inducible transcript 3 (DDIT-3) (Growth arrest and DNA- damage-inducible protein GADD153) (C/EBP-homologous protein) (CHOP). | |||||
|
GCC2_MOUSE
|
||||||
| NC score | 0.054655 (rank : 18) | θ value | 0.125558 (rank : 19) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1619 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q8CHG3, Q8BR44, Q8R2Q5, Q9CT45 | Gene names | Gcc2, Kiaa0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185). | |||||
|
CENPF_HUMAN
|
||||||
| NC score | 0.053822 (rank : 19) | θ value | 0.125558 (rank : 18) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1932 | Shared Neighborhood Hits | 63 | |
| SwissProt Accessions | P49454, Q13171, Q13246 | Gene names | CENPF | |||
|
Domain Architecture |

|
|||||
| Description | Centromere protein F (Kinetochore protein CENP-F) (Mitosin) (AH antigen). | |||||
|
CF060_HUMAN
|
||||||
| NC score | 0.053702 (rank : 20) | θ value | 0.0961366 (rank : 17) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1270 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q8NB25, Q96GY8, Q9H851 | Gene names | C6orf60 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Uncharacterized protein C6orf60. | |||||
|
CP250_MOUSE
|
||||||
| NC score | 0.052600 (rank : 21) | θ value | 0.0431538 (rank : 15) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1583 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q60952, Q2I8G3, Q3UTR4, Q6PFF6, Q8BLC6 | Gene names | Cep250, Cep2, Inmp | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Centrosome-associated protein CEP250 (Centrosomal protein 2) (Centrosomal Nek2-associated protein 1) (C-Nap1) (Intranuclear matrix protein). | |||||
|
MYOC_HUMAN
|
||||||
| NC score | 0.052333 (rank : 22) | θ value | 0.0736092 (rank : 16) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 489 | Shared Neighborhood Hits | 37 | |
| SwissProt Accessions | Q99972, O00620, Q7Z6Q9 | Gene names | MYOC, GLC1A, TIGR | |||
|
Domain Architecture |

|
|||||
| Description | Myocilin precursor (Trabecular meshwork-induced glucocorticoid response protein). | |||||
|
PCNT_HUMAN
|
||||||
| NC score | 0.051779 (rank : 23) | θ value | 4.03905 (rank : 62) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1551 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | O95613, O43152, Q7Z7C9 | Gene names | PCNT, KIAA0402, PCNT2 | |||
|
Domain Architecture |

|
|||||
| Description | Pericentrin (Pericentrin B) (Kendrin). | |||||
|
CROCC_HUMAN
|
||||||
| NC score | 0.051659 (rank : 24) | θ value | 2.36792 (rank : 46) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1409 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | Q5TZA2, Q2VHY3, Q66GT7, Q7Z2L4, Q7Z5D7 | Gene names | CROCC, KIAA0445 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rootletin (Ciliary rootlet coiled-coil protein). | |||||
|
RBP24_HUMAN
|
||||||
| NC score | 0.051469 (rank : 25) | θ value | 0.21417 (rank : 23) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1559 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q4ZG46 | Gene names | RANBP2L4 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ran-binding protein 2-like 4 (RanBP2L4). | |||||
|
GCC2_HUMAN
|
||||||
| NC score | 0.051374 (rank : 26) | θ value | 0.21417 (rank : 22) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1558 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | Q8IWJ2, O15045, Q8TDH3, Q9H2G8 | Gene names | GCC2, KIAA0336 | |||
|
Domain Architecture |

|
|||||
| Description | GRIP and coiled-coil domain-containing protein 2 (Golgi coiled coil protein GCC185) (CTCL tumor antigen se1-1) (CLL-associated antigen KW- 11) (NY-REN-53 antigen). | |||||
|
MYH9_MOUSE
|
||||||
| NC score | 0.044769 (rank : 27) | θ value | 0.125558 (rank : 20) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1829 | Shared Neighborhood Hits | 64 | |
| SwissProt Accessions | Q8VDD5 | Gene names | Myh9 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
MYH9_HUMAN
|
||||||
| NC score | 0.043279 (rank : 28) | θ value | 0.163984 (rank : 21) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1762 | Shared Neighborhood Hits | 62 | |
| SwissProt Accessions | P35579, O60805 | Gene names | MYH9 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A). | |||||
|
BICD2_HUMAN
|
||||||
| NC score | 0.042802 (rank : 29) | θ value | 0.813845 (rank : 29) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q8TD16, O75181, Q5TBQ2, Q5TBQ3, Q96LH2, Q9BT84, Q9H561 | Gene names | BICD2, KIAA0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
BICD2_MOUSE
|
||||||
| NC score | 0.042668 (rank : 30) | θ value | 2.36792 (rank : 45) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 864 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q921C5, Q80TU1, Q8BTE3, Q9DCL3 | Gene names | Bicd2, Kiaa0699 | |||
|
Domain Architecture |

|
|||||
| Description | Cytoskeleton-like bicaudal D protein homolog 2. | |||||
|
GOGA2_HUMAN
|
||||||
| NC score | 0.042445 (rank : 31) | θ value | 4.03905 (rank : 59) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 856 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | Q08379, Q9BRB0, Q9NYF9 | Gene names | GOLGA2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Golgin subfamily A member 2 (Cis-Golgi matrix protein GM130) (Gm130 autoantigen) (Golgin-95). | |||||
|
FLIP1_MOUSE
|
||||||
| NC score | 0.042036 (rank : 32) | θ value | 5.27518 (rank : 65) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1176 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q9CS72 | Gene names | Filip1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
MYH11_HUMAN
|
||||||
| NC score | 0.041609 (rank : 33) | θ value | 0.365318 (rank : 26) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1806 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | P35749, O00396, O94944, P78422 | Gene names | MYH11, KIAA0866 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
CCD39_HUMAN
|
||||||
| NC score | 0.041317 (rank : 34) | θ value | 4.03905 (rank : 56) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 868 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9UFE4 | Gene names | CCDC39 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled-coil domain-containing protein 39. | |||||
|
CING_MOUSE
|
||||||
| NC score | 0.040953 (rank : 35) | θ value | 8.99809 (rank : 81) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1432 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P59242 | Gene names | Cgn | |||
|
Domain Architecture |

|
|||||
| Description | Cingulin. | |||||
|
SAS6_MOUSE
|
||||||
| NC score | 0.040446 (rank : 36) | θ value | 0.813845 (rank : 32) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 931 | Shared Neighborhood Hits | 51 | |
| SwissProt Accessions | Q80UK7, Q9CYT4 | Gene names | Sass6, Sas6 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Spindle assembly abnormal protein 6 homolog. | |||||
|
CCD46_MOUSE
|
||||||
| NC score | 0.040179 (rank : 37) | θ value | 5.27518 (rank : 64) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q5PR68, Q80ZN6, Q99JS4, Q9D9T1 | Gene names | Ccdc46 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Coiled coil domain-containing protein 46. | |||||
|
LRRF2_MOUSE
|
||||||
| NC score | 0.039586 (rank : 38) | θ value | 5.27518 (rank : 67) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 556 | Shared Neighborhood Hits | 39 | |
| SwissProt Accessions | Q91WK0, Q8BVD1, Q9CU89 | Gene names | Lrrfip2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 2 (LRR FLII- interacting protein 2). | |||||
|
FLIP1_HUMAN
|
||||||
| NC score | 0.039249 (rank : 39) | θ value | 6.88961 (rank : 71) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1405 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | Q7Z7B0, Q5VUL6, Q86TC3, Q8N8B9, Q96SK6, Q9NVI8, Q9ULE5 | Gene names | FILIP1, KIAA1275 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Filamin-A-interacting protein 1 (FILIP). | |||||
|
RB6I2_HUMAN
|
||||||
| NC score | 0.038865 (rank : 40) | θ value | 6.88961 (rank : 76) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1584 | Shared Neighborhood Hits | 58 | |
| SwissProt Accessions | Q8IUD2, Q6NVK2, Q8IUD3, Q8IUD4, Q8IUD5, Q8NAS1, Q9NXN5, Q9UIK7, Q9UPS1 | Gene names | ERC1, ELKS, KIAA1081, RAB6IP2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ELKS/RAB6-interacting/CAST family member 1 (RAB6-interacting protein 2) (ERC protein 1). | |||||
|
MYH11_MOUSE
|
||||||
| NC score | 0.038708 (rank : 41) | θ value | 6.88961 (rank : 75) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1777 | Shared Neighborhood Hits | 59 | |
| SwissProt Accessions | O08638, O08639, Q62462, Q64195 | Gene names | Myh11 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC). | |||||
|
REST_MOUSE
|
||||||
| NC score | 0.038478 (rank : 42) | θ value | 8.99809 (rank : 84) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1506 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q922J3, Q571L7 | Gene names | Rsn, Kiaa4046 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Restin. | |||||
|
BPA1_MOUSE
|
||||||
| NC score | 0.038418 (rank : 43) | θ value | 0.365318 (rank : 25) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1477 | Shared Neighborhood Hits | 48 | |
| SwissProt Accessions | Q91ZU6, Q91ZU7 | Gene names | Dst, Bpag1 | |||
|
Domain Architecture |

|
|||||
| Description | Bullous pemphigoid antigen 1, isoforms 1/2/3/4 (BPA) (Hemidesmosomal plaque protein) (Dystonia musculorum protein) (Dystonin). | |||||
|
MACF1_MOUSE
|
||||||
| NC score | 0.037645 (rank : 44) | θ value | 0.813845 (rank : 31) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1313 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | Q9QXZ0, P97394, P97395, P97396 | Gene names | Macf1, Acf7, Aclp7, Macf | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1 (Actin cross-linking family 7). | |||||
|
EMIL2_MOUSE
|
||||||
| NC score | 0.037284 (rank : 45) | θ value | 1.38821 (rank : 41) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 485 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q8K482 | Gene names | Emilin2 | |||
|
Domain Architecture |

|
|||||
| Description | EMILIN-2 precursor (Elastin microfibril interface-located protein 2) (Elastin microfibril interfacer 2) (Basilin). | |||||
|
GFAP_HUMAN
|
||||||
| NC score | 0.035818 (rank : 46) | θ value | 0.62314 (rank : 28) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 723 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P14136, Q53H98, Q5D055, Q6ZQS3, Q7Z5J6, Q7Z5J7, Q96KS4, Q96P18, Q9UFD0 | Gene names | GFAP | |||
|
Domain Architecture |

|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
IFT81_MOUSE
|
||||||
| NC score | 0.035745 (rank : 47) | θ value | 4.03905 (rank : 60) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 959 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | O35594, Q8R4W2, Q9CSA1, Q9EQM1 | Gene names | Ift81, Cdv-1, Cdv1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Intraflagellar transport 81 (Carnitine deficiency-associated protein expressed in ventricle 1) (CDV-1 protein). | |||||
|
CENPE_HUMAN
|
||||||
| NC score | 0.034517 (rank : 48) | θ value | 4.03905 (rank : 57) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 2060 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | Q02224 | Gene names | CENPE | |||
|
Domain Architecture |

|
|||||
| Description | Centromeric protein E (CENP-E). | |||||
|
CASP_MOUSE
|
||||||
| NC score | 0.034194 (rank : 49) | θ value | 6.88961 (rank : 69) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1152 | Shared Neighborhood Hits | 53 | |
| SwissProt Accessions | P70403, Q7TQJ4, Q91WN6 | Gene names | Cutl1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein CASP. | |||||
|
MYH8_HUMAN
|
||||||
| NC score | 0.033980 (rank : 50) | θ value | 2.36792 (rank : 48) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1610 | Shared Neighborhood Hits | 57 | |
| SwissProt Accessions | P13535, Q14910 | Gene names | MYH8 | |||
|
Domain Architecture |

|
|||||
| Description | Myosin-8 (Myosin heavy chain, skeletal muscle, perinatal) (MyHC- perinatal). | |||||
|
GFAP_MOUSE
|
||||||
| NC score | 0.033420 (rank : 51) | θ value | 1.81305 (rank : 43) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 725 | Shared Neighborhood Hits | 49 | |
| SwissProt Accessions | P03995, Q7TQ30, Q925K3 | Gene names | Gfap | |||
|
Domain Architecture |

|
|||||
| Description | Glial fibrillary acidic protein, astrocyte (GFAP). | |||||
|
CUTL1_MOUSE
|
||||||
| NC score | 0.031554 (rank : 52) | θ value | 3.0926 (rank : 51) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1620 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P53564, O08994, P70301, Q571L6, Q91ZD2 | Gene names | Cutl1, Cux, Cux1, Kiaa4047 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Homeobox protein cut-like 1 (CCAAT displacement protein) (CDP) (Homeobox protein Cux). | |||||
|
RIMB1_MOUSE
|
||||||
| NC score | 0.031155 (rank : 53) | θ value | 8.99809 (rank : 85) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1376 | Shared Neighborhood Hits | 50 | |
| SwissProt Accessions | Q7TNF8, Q5NCP7, Q80TV9, Q8BIH5 | Gene names | Bzrap1, Kiaa0612, Rbp1 | |||
|
Domain Architecture |

|
|||||
| Description | Peripheral-type benzodiazepine receptor-associated protein 1 (PRAX-1) (Peripheral benzodiazepine receptor-interacting protein) (PBR-IP) (RIM-binding protein 1) (RIM-BP1). | |||||
|
KINH_HUMAN
|
||||||
| NC score | 0.030859 (rank : 54) | θ value | 1.81305 (rank : 44) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1231 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | P33176, Q5VZ85 | Gene names | KIF5B, KNS, KNS1 | |||
|
Domain Architecture |

|
|||||
| Description | Kinesin heavy chain (Ubiquitous kinesin heavy chain) (UKHC). | |||||
|
SPTN5_HUMAN
|
||||||
| NC score | 0.030368 (rank : 55) | θ value | 3.0926 (rank : 55) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 674 | Shared Neighborhood Hits | 44 | |
| SwissProt Accessions | Q9NRC6 | Gene names | SPTBN5, SPTBN4 | |||
|
Domain Architecture |

|
|||||
| Description | Spectrin beta chain, brain 4 (Spectrin, non-erythroid beta chain 4) (Beta-V spectrin) (BSPECV). | |||||
|
TPD54_HUMAN
|
||||||
| NC score | 0.030304 (rank : 56) | θ value | 4.03905 (rank : 63) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O43399, Q9H3Z6 | Gene names | TPD52L2 | |||
|
Domain Architecture |

|
|||||
| Description | Tumor protein D54 (hD54) (Tumor protein D52-like 2). | |||||
|
KRHB6_HUMAN
|
||||||
| NC score | 0.028873 (rank : 57) | θ value | 1.06291 (rank : 39) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 447 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | O43790, P78387 | Gene names | KRTHB6 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb6 (Hair keratin, type II Hb6) (ghHb6). | |||||
|
KRHB1_HUMAN
|
||||||
| NC score | 0.028753 (rank : 58) | θ value | 1.06291 (rank : 37) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 461 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | Q14533, Q14846, Q16274, Q8WU52, Q9BR74 | Gene names | KRTHB1, MLN137 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb1 (Hair keratin, type II Hb1) (ghHKb1) (ghHb1) (MLN 137). | |||||
|
TRI32_MOUSE
|
||||||
| NC score | 0.028662 (rank : 59) | θ value | 0.365318 (rank : 27) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 271 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | Q8CH72, Q8K055 | Gene names | Trim32 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-). | |||||
|
KRHB3_HUMAN
|
||||||
| NC score | 0.028550 (rank : 60) | θ value | 1.06291 (rank : 38) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 438 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P78385, Q9NSB3 | Gene names | KRTHB3 | |||
|
Domain Architecture |

|
|||||
| Description | Keratin, type II cuticular Hb3 (Hair keratin, type II Hb3). | |||||
|
LRRF1_HUMAN
|
||||||
| NC score | 0.027013 (rank : 61) | θ value | 6.88961 (rank : 74) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 625 | Shared Neighborhood Hits | 31 | |
| SwissProt Accessions | Q32MZ4, O75766, O75799, Q32MZ5, Q53T49, Q6PKG2 | Gene names | LRRFIP1, GCF2, TRIP | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Leucine-rich repeat flightless-interacting protein 1 (LRR FLII- interacting protein 1) (TAR RNA-interacting protein) (GC-binding factor 2). | |||||
|
KRHB6_MOUSE
|
||||||
| NC score | 0.026658 (rank : 62) | θ value | 2.36792 (rank : 47) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 413 | Shared Neighborhood Hits | 35 | |
| SwissProt Accessions | P97861, Q3TTV9 | Gene names | Krthb6, Krt2-10 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Keratin, type II cuticular Hb6 (Hair keratin, type II Hb6). | |||||
|
NFH_HUMAN
|
||||||
| NC score | 0.025717 (rank : 63) | θ value | 3.0926 (rank : 54) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1242 | Shared Neighborhood Hits | 47 | |
| SwissProt Accessions | P12036, Q9UJS7, Q9UQ14 | Gene names | NEFH, KIAA0845, NFH | |||
|
Domain Architecture |

|
|||||
| Description | Neurofilament triplet H protein (200 kDa neurofilament protein) (Neurofilament heavy polypeptide) (NF-H). | |||||
|
GLE1_HUMAN
|
||||||
| NC score | 0.025183 (rank : 64) | θ value | 6.88961 (rank : 72) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 445 | Shared Neighborhood Hits | 27 | |
| SwissProt Accessions | Q53GS7, O75458, Q53GT9, Q5VVU1, Q8NCP6, Q9UFL6 | Gene names | GLE1L, GLE1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleoporin GLE1 (GLE1-like protein) (hGLE1). | |||||
|
ANR35_HUMAN
|
||||||
| NC score | 0.022500 (rank : 65) | θ value | 1.06291 (rank : 34) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 784 | Shared Neighborhood Hits | 43 | |
| SwissProt Accessions | Q8N283, Q3MJ10, Q96LS3 | Gene names | ANKRD35 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ankyrin repeat domain-containing protein 35. | |||||
|
TRIM3_HUMAN
|
||||||
| NC score | 0.022406 (rank : 66) | θ value | 0.813845 (rank : 33) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 357 | Shared Neighborhood Hits | 15 | |
| SwissProt Accessions | O75382, Q9C038, Q9C039 | Gene names | TRIM3, BERP, RNF22, RNF97 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 3 (RING finger protein 22) (Brain- expressed RING finger protein) (RING finger protein 97). | |||||
|
NUCB2_MOUSE
|
||||||
| NC score | 0.022388 (rank : 67) | θ value | 8.99809 (rank : 83) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 459 | Shared Neighborhood Hits | 34 | |
| SwissProt Accessions | P81117 | Gene names | Nucb2, Nefa | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Nucleobindin-2 precursor (DNA-binding protein NEFA). | |||||
|
TRI32_HUMAN
|
||||||
| NC score | 0.021698 (rank : 68) | θ value | 6.88961 (rank : 78) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 264 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q13049, Q9NQP8 | Gene names | TRIM32, HT2A | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 32 (EC 6.3.2.-) (Zinc finger protein HT2A) (72 kDa Tat-interacting protein). | |||||
|
CD72_HUMAN
|
||||||
| NC score | 0.020542 (rank : 69) | θ value | 8.99809 (rank : 80) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 345 | Shared Neighborhood Hits | 32 | |
| SwissProt Accessions | P21854 | Gene names | CD72 | |||
|
Domain Architecture |

|
|||||
| Description | B-cell differentiation antigen CD72 (Lyb-2). | |||||
|
FNBP2_HUMAN
|
||||||
| NC score | 0.020095 (rank : 70) | θ value | 5.27518 (rank : 66) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 432 | Shared Neighborhood Hits | 25 | |
| SwissProt Accessions | O75044 | Gene names | SRGAP2, FNBP2, KIAA0456 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 2 (srGAP2) (Formin-binding protein 2). | |||||
|
CGRE1_HUMAN
|
||||||
| NC score | 0.019178 (rank : 71) | θ value | 1.38821 (rank : 40) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 354 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99674 | Gene names | CGREF1, CGR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Cell growth regulator with EF hand domain 1 (Cell growth regulatory gene 11 protein). | |||||
|
SRGP2_MOUSE
|
||||||
| NC score | 0.018729 (rank : 72) | θ value | 2.36792 (rank : 49) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 324 | Shared Neighborhood Hits | 21 | |
| SwissProt Accessions | Q812A2, Q80U09, Q8BKP4, Q8BLD0, Q925I2 | Gene names | Srgap3, Arhgap14, Kiaa0411, Srgap2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Rho-GTPase-activating protein 14). | |||||
|
TRIM3_MOUSE
|
||||||
| NC score | 0.017788 (rank : 73) | θ value | 2.36792 (rank : 50) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 351 | Shared Neighborhood Hits | 13 | |
| SwissProt Accessions | Q9R1R2, Q3UMU5 | Gene names | Trim3, Hac1, Rnf22 | |||
|
Domain Architecture |

|
|||||
| Description | Tripartite motif-containing protein 3 (RING finger protein 22) (RING finger protein HAC1). | |||||
|
ROCK2_MOUSE
|
||||||
| NC score | 0.017728 (rank : 74) | θ value | 5.27518 (rank : 68) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 2072 | Shared Neighborhood Hits | 52 | |
| SwissProt Accessions | P70336, Q8BR64, Q8CBR0 | Gene names | Rock2 | |||
|
Domain Architecture |

|
|||||
| Description | Rho-associated protein kinase 2 (EC 2.7.11.1) (Rho-associated, coiled- coil-containing protein kinase 2) (p164 ROCK-2). | |||||
|
DOCK6_HUMAN
|
||||||
| NC score | 0.016882 (rank : 75) | θ value | 1.81305 (rank : 42) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 71 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q96HP0, Q9P2F2 | Gene names | DOCK6, KIAA1395 | |||
|
Domain Architecture |

|
|||||
| Description | Dedicator of cytokinesis protein 6. | |||||
|
SRGP2_HUMAN
|
||||||
| NC score | 0.015754 (rank : 76) | θ value | 8.99809 (rank : 86) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 331 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O43295, Q8IX13, Q8IZV8 | Gene names | SRGAP3, ARHGAP14, KIAA0411, MEGAP, SRGAP2 | |||
|
Domain Architecture |

|
|||||
| Description | SLIT-ROBO Rho GTPase-activating protein 3 (srGAP3) (srGAP2) (WAVE- associated Rac GTPase-activating protein) (WRP) (Mental disorder- associated GAP) (Rho-GTPase-activating protein 14). | |||||
|
MRCKG_HUMAN
|
||||||
| NC score | 0.012524 (rank : 77) | θ value | 3.0926 (rank : 53) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1519 | Shared Neighborhood Hits | 54 | |
| SwissProt Accessions | Q6DT37, O00565 | Gene names | CDC42BPG | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Serine/threonine-protein kinase MRCK gamma (EC 2.7.11.1) (CDC42- binding protein kinase gamma) (Myotonic dystrophy kinase-related CDC42-binding kinase gamma) (Myotonic dystrophy protein kinase-like alpha) (MRCK gamma) (DMPK-like gamma). | |||||
|
CNTP4_HUMAN
|
||||||
| NC score | 0.011588 (rank : 78) | θ value | 4.03905 (rank : 58) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 187 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9C0A0 | Gene names | CNTNAP4, CASPR4, KIAA1763 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
APXL_HUMAN
|
||||||
| NC score | 0.011525 (rank : 79) | θ value | 8.99809 (rank : 79) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 553 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q13796 | Gene names | APXL | |||
|
Domain Architecture |

|
|||||
| Description | Apical-like protein (Protein APXL). | |||||
|
FARP2_MOUSE
|
||||||
| NC score | 0.011381 (rank : 80) | θ value | 0.813845 (rank : 30) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q91VS8, Q3TAP2, Q69ZZ0 | Gene names | Farp2, Kiaa0793 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FERM, RhoGEF and pleckstrin domain-containing protein 2 (FERM domain including RhoGEF) (FIR). | |||||
|
LAMB2_MOUSE
|
||||||
| NC score | 0.010303 (rank : 81) | θ value | 6.88961 (rank : 73) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 722 | Shared Neighborhood Hits | 36 | |
| SwissProt Accessions | Q61292, Q62182 | Gene names | Lamb2, Lams | |||
|
Domain Architecture |

|
|||||
| Description | Laminin beta-2 chain precursor (S-laminin) (S-LAM). | |||||
|
CNTP4_MOUSE
|
||||||
| NC score | 0.009929 (rank : 82) | θ value | 6.88961 (rank : 70) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 193 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q99P47 | Gene names | Cntnap4, Caspr4 | |||
|
Domain Architecture |

|
|||||
| Description | Contactin-associated protein-like 4 precursor (Cell recognition molecule Caspr4). | |||||
|
KGP2_HUMAN
|
||||||
| NC score | 0.005437 (rank : 83) | θ value | 1.06291 (rank : 36) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1023 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q13237, O00125, O60916 | Gene names | PRKG2, PRKGR2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
MATN4_HUMAN
|
||||||
| NC score | 0.004250 (rank : 84) | θ value | 3.0926 (rank : 52) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 287 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | O95460, Q5QPU2, Q5QPU3, Q5QPU4, Q8N2M5, Q8N2M7, Q9H1F8, Q9H1F9 | Gene names | MATN4 | |||
|
Domain Architecture |

|
|||||
| Description | Matrilin-4 precursor. | |||||
|
KGP2_MOUSE
|
||||||
| NC score | 0.004024 (rank : 85) | θ value | 4.03905 (rank : 61) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 1029 | Shared Neighborhood Hits | 26 | |
| SwissProt Accessions | Q61410 | Gene names | Prkg2, Prkgr2 | |||
|
Domain Architecture |

|
|||||
| Description | cGMP-dependent protein kinase 2 (EC 2.7.11.12) (CGK 2) (cGKII) (Type II cGMP-dependent protein kinase). | |||||
|
COBA2_MOUSE
|
||||||
| NC score | 0.001943 (rank : 86) | θ value | 8.99809 (rank : 82) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 501 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q64739, Q61432, Q9Z1W0 | Gene names | Col11a2 | |||
|
Domain Architecture |

|
|||||
| Description | Collagen alpha-2(XI) chain precursor. | |||||
|
TENA_HUMAN
|
||||||
| NC score | 0.001870 (rank : 87) | θ value | 6.88961 (rank : 77) | |||
| Query Neighborhood Hits | 86 | Target Neighborhood Hits | 631 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P24821, Q14583, Q15567 | Gene names | TNC, HXB | |||
|
Domain Architecture |

|
|||||
| Description | Tenascin precursor (TN) (Tenascin-C) (TN-C) (Hexabrachion) (Cytotactin) (Neuronectin) (GMEM) (JI) (Myotendinous antigen) (Glioma- associated-extracellular matrix antigen) (GP 150-225). | |||||