Please be patient as the page loads
|
MUCM_HUMAN
|
||||||
| SwissProt Accessions | P20769 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig mu chain C region membrane-bound segment. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MUCM_HUMAN
|
||||||
| θ value | 6.85773e-16 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20769 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig mu chain C region membrane-bound segment. | |||||
|
MUCM_MOUSE
|
||||||
| θ value | 1.29331e-14 (rank : 2) | NC score | 0.413648 (rank : 2) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01873 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig mu chain C region membrane-bound form. | |||||
|
GC3M_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 3) | NC score | 0.348744 (rank : 4) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P03987 | Gene names | ||||
|
Domain Architecture |
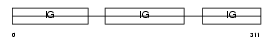
|
|||||
| Description | Ig gamma-3 chain C region, membrane-bound form. | |||||
|
GCAM_MOUSE
|
||||||
| θ value | 0.00175202 (rank : 4) | NC score | 0.352698 (rank : 3) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01865 | Gene names | Igh-1a | |||
|
Domain Architecture |
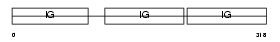
|
|||||
| Description | Ig gamma-2A chain C region, membrane-bound form. | |||||
|
GCBM_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 5) | NC score | 0.348141 (rank : 5) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01867 | Gene names | ||||
|
Domain Architecture |
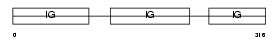
|
|||||
| Description | Ig gamma-2B chain C region, membrane-bound form. | |||||
|
IGH1M_MOUSE
|
||||||
| θ value | 0.00228821 (rank : 6) | NC score | 0.345139 (rank : 6) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01869 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig gamma-1 chain C region, membrane-bound form. | |||||
|
IGHDM_MOUSE
|
||||||
| θ value | 0.0252991 (rank : 7) | NC score | 0.323745 (rank : 8) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01882 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig delta chain C region membrane-bound form. | |||||
|
2DMB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 8) | NC score | 0.057143 (rank : 45) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P28068, O77936, Q13012, Q29751, Q9XRX2 | Gene names | HLA-DMB, DMB, RING7 | |||
|
Domain Architecture |
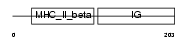
|
|||||
| Description | HLA class II histocompatibility antigen, DM beta chain precursor (MHC class II antigen DMB). | |||||
|
2DMB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 9) | NC score | 0.068926 (rank : 43) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35737 | Gene names | H2-DMb1, H-2Mb1, Mb | |||
|
Domain Architecture |
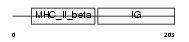
|
|||||
| Description | Class II histocompatibility antigen, M beta 1 chain precursor (H2-M beta 1 chain). | |||||
|
GC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 10) | NC score | 0.296321 (rank : 15) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22436 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig gamma-3 chain C region, secreted form. | |||||
|
GCAA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 11) | NC score | 0.302574 (rank : 10) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01863 | Gene names | ||||
|
Domain Architecture |
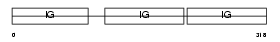
|
|||||
| Description | Ig gamma-2A chain C region, A allele. | |||||
|
GCAB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 12) | NC score | 0.300809 (rank : 11) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01864 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig gamma-2A chain C region secreted form (B allele). | |||||
|
GCB_MOUSE
|
||||||
| θ value | θ > 10 (rank : 13) | NC score | 0.299594 (rank : 13) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01866 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig gamma-2B chain C region secreted form. | |||||
|
IGHA1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 14) | NC score | 0.266453 (rank : 23) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01876 | Gene names | IGHA1 | |||
|
Domain Architecture |
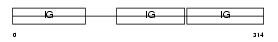
|
|||||
| Description | Ig alpha-1 chain C region. | |||||
|
IGHA2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 15) | NC score | 0.267282 (rank : 22) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01877 | Gene names | IGHA2 | |||
|
Domain Architecture |
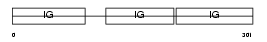
|
|||||
| Description | Ig alpha-2 chain C region. | |||||
|
IGHA_MOUSE
|
||||||
| θ value | θ > 10 (rank : 16) | NC score | 0.257409 (rank : 25) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01878 | Gene names | ||||
|
Domain Architecture |
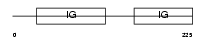
|
|||||
| Description | Ig alpha chain C region. | |||||
|
IGHD_HUMAN
|
||||||
| θ value | θ > 10 (rank : 17) | NC score | 0.268298 (rank : 21) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01880 | Gene names | IGHD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig delta chain C region. | |||||
|
IGHD_MOUSE
|
||||||
| θ value | θ > 10 (rank : 18) | NC score | 0.191438 (rank : 26) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01881 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig delta chain C region secreted form. | |||||
|
IGHE_HUMAN
|
||||||
| θ value | θ > 10 (rank : 19) | NC score | 0.293042 (rank : 16) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01854 | Gene names | IGHE | |||
|
Domain Architecture |

|
|||||
| Description | Ig epsilon chain C region. | |||||
|
IGHE_MOUSE
|
||||||
| θ value | θ > 10 (rank : 20) | NC score | 0.263871 (rank : 24) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06336, P01856 | Gene names | ||||
|
Domain Architecture |
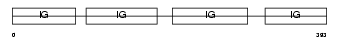
|
|||||
| Description | Ig epsilon chain C region. | |||||
|
IGHG1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 21) | NC score | 0.285387 (rank : 19) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01857 | Gene names | IGHG1 | |||
|
Domain Architecture |
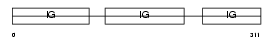
|
|||||
| Description | Ig gamma-1 chain C region. | |||||
|
IGHG1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 22) | NC score | 0.298803 (rank : 14) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01868 | Gene names | ||||
|
Domain Architecture |
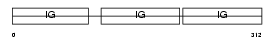
|
|||||
| Description | Ig gamma-1 chain C region secreted form. | |||||
|
IGHG2_HUMAN
|
||||||
| θ value | θ > 10 (rank : 23) | NC score | 0.287288 (rank : 18) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01859 | Gene names | IGHG2 | |||
|
Domain Architecture |
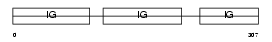
|
|||||
| Description | Ig gamma-2 chain C region. | |||||
|
IGHG3_HUMAN
|
||||||
| θ value | θ > 10 (rank : 24) | NC score | 0.279376 (rank : 20) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01860 | Gene names | IGHG3 | |||
|
Domain Architecture |
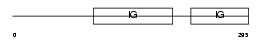
|
|||||
| Description | Ig gamma-3 chain C region (Heavy chain disease protein) (HDC). | |||||
|
IGHG4_HUMAN
|
||||||
| θ value | θ > 10 (rank : 25) | NC score | 0.287315 (rank : 17) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01861 | Gene names | IGHG4 | |||
|
Domain Architecture |
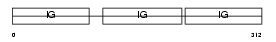
|
|||||
| Description | Ig gamma-4 chain C region. | |||||
|
IGLL1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 26) | NC score | 0.138296 (rank : 34) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P15814 | Gene names | IGLL1, IGL1 | |||
|
Domain Architecture |
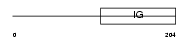
|
|||||
| Description | Immunoglobulin lambda-like polypeptide 1 precursor (Immunoglobulin- related protein 14.1) (Immunoglobulin omega polypeptide) (Ig lambda-5) (CD179b antigen). | |||||
|
IGLL1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 27) | NC score | 0.140157 (rank : 33) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20764, Q5W1K3 | Gene names | Igll1, Igl-5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin lambda-like polypeptide 1 precursor (Ig lambda-5) (CD179b antigen). | |||||
|
KAC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 28) | NC score | 0.173055 (rank : 27) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01834 | Gene names | IGKC | |||
|
Domain Architecture |
|
|||||
| Description | Ig kappa chain C region. | |||||
|
KAC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 29) | NC score | 0.164915 (rank : 29) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01837 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Ig kappa chain C region. | |||||
|
LAC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 30) | NC score | 0.153085 (rank : 31) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01843 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Ig lambda-1 chain C region. | |||||
|
LAC2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 31) | NC score | 0.164878 (rank : 30) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01844 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Ig lambda-2 chain C region. | |||||
|
LAC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 32) | NC score | 0.172649 (rank : 28) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01845 | Gene names | ||||
|
Domain Architecture |
|
|||||
| Description | Ig lambda-3 chain C region. | |||||
|
LAC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 33) | NC score | 0.152849 (rank : 32) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01842, P80423 | Gene names | IGLC1 | |||
|
Domain Architecture |
|
|||||
| Description | Ig lambda chain C regions. | |||||
|
MUCB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 34) | NC score | 0.300663 (rank : 12) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P04220 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig mu heavy chain disease protein (BOT). | |||||
|
MUC_HUMAN
|
||||||
| θ value | θ > 10 (rank : 35) | NC score | 0.307039 (rank : 9) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01871 | Gene names | IGHM | |||
|
Domain Architecture |

|
|||||
| Description | Ig mu chain C region. | |||||
|
MUC_MOUSE
|
||||||
| θ value | θ > 10 (rank : 36) | NC score | 0.327894 (rank : 7) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01872 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig mu chain C region secreted form. | |||||
|
SHPS1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 37) | NC score | 0.080293 (rank : 41) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78324, O00683, O43799, Q8N517, Q8TAL8, Q9H0Z2, Q9UDX2, Q9UIJ6, Q9Y4U9 | Gene names | SIRPA, BIT, MFR, MYD1, PTPNS1, SHPS1, SIRP | |||
|
Domain Architecture |
|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type substrate 1 precursor (SHP substrate 1) (SHPS-1) (Inhibitory receptor SHPS-1) (Signal- regulatory protein alpha-1) (Sirp-alpha-1) (Sirp-alpha-2) (Sirp-alpha- 3) (MyD-1 antigen) (Brain Ig-like molecule with tyrosine-based activation motifs) (Bit) (Macrophage fusion receptor) (p84) (CD172a antigen). | |||||
|
SHPS1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 38) | NC score | 0.101570 (rank : 39) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97797, O08907, O35924, O88555, O88556, P97796, Q8R559, Q9QX57, Q9WTN4 | Gene names | Sirpa, Bit, Myd1, Ptpns1, Shps1, Sirp | |||
|
Domain Architecture |
|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type substrate 1 precursor (SHP substrate 1) (SHPS-1) (Inhibitory receptor SHPS-1) (Signal- regulatory protein alpha-1) (Sirp-alpha-1) (mSIRP-alpha1) (MyD-1 antigen) (Brain Ig-like molecule with tyrosine-based activation motifs) (Bit) (p84) (CD172a antigen). | |||||
|
SIRB1_HUMAN
|
||||||
| θ value | θ > 10 (rank : 39) | NC score | 0.086822 (rank : 40) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00241, Q5TFR0, Q8TB12, Q9H1U5, Q9Y4V0 | Gene names | SIRPB1 | |||
|
Domain Architecture |
|
|||||
| Description | Signal-regulatory protein beta-1 precursor (SIRP-beta-1) (CD172b antigen). | |||||
|
SIRPG_HUMAN
|
||||||
| θ value | θ > 10 (rank : 40) | NC score | 0.102323 (rank : 38) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P1W8, Q8WWA5, Q9NQK8 | Gene names | SIRPG, SIRPB2 | |||
|
Domain Architecture |
|
|||||
| Description | Signal-regulatory protein gamma precursor (Signal-regulatory protein beta-2) (SIRP-beta-2) (SIRP-b2) (CD172g antigen). | |||||
|
TCB1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 41) | NC score | 0.131171 (rank : 37) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01852 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell receptor beta-1 chain C region. | |||||
|
TCB2_MOUSE
|
||||||
| θ value | θ > 10 (rank : 42) | NC score | 0.133383 (rank : 36) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01851 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell receptor beta-2 chain C region. | |||||
|
TCB_HUMAN
|
||||||
| θ value | θ > 10 (rank : 43) | NC score | 0.134823 (rank : 35) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01850 | Gene names | TRBC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell receptor beta chain C region. | |||||
|
TCC1_MOUSE
|
||||||
| θ value | θ > 10 (rank : 44) | NC score | 0.072423 (rank : 42) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01853 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell receptor gamma chain C region C10.5. | |||||
|
TCC3_MOUSE
|
||||||
| θ value | θ > 10 (rank : 45) | NC score | 0.051658 (rank : 47) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P06334 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell receptor gamma chain C region DFL12. | |||||
|
TCC4_MOUSE
|
||||||
| θ value | θ > 10 (rank : 46) | NC score | 0.055703 (rank : 46) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06335 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell receptor gamma chain C region 5/10-13. | |||||
|
TPSNR_HUMAN
|
||||||
| θ value | θ > 10 (rank : 47) | NC score | 0.050998 (rank : 48) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BX59, Q9NWB8 | Gene names | TAPBPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tapasin-related protein precursor (TAPASIN-R) (Tapasin-like) (TAP- binding protein-related protein) (TAPBP-R) (TAP-binding protein-like). | |||||
|
TPSNR_MOUSE
|
||||||
| θ value | θ > 10 (rank : 48) | NC score | 0.063137 (rank : 44) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VD31 | Gene names | Tapbpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tapasin-related protein precursor (TAPASIN-R) (Tapasin-like) (TAP- binding protein-related protein) (TAPBP-R) (TAP-binding protein-like). | |||||
|
MUCM_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 6.85773e-16 (rank : 1) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P20769 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig mu chain C region membrane-bound segment. | |||||
|
MUCM_MOUSE
|
||||||
| NC score | 0.413648 (rank : 2) | θ value | 1.29331e-14 (rank : 2) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01873 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig mu chain C region membrane-bound form. | |||||
|
GCAM_MOUSE
|
||||||
| NC score | 0.352698 (rank : 3) | θ value | 0.00175202 (rank : 4) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01865 | Gene names | Igh-1a | |||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-2A chain C region, membrane-bound form. | |||||
|
GC3M_MOUSE
|
||||||
| NC score | 0.348744 (rank : 4) | θ value | 0.00175202 (rank : 3) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 215 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P03987 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-3 chain C region, membrane-bound form. | |||||
|
GCBM_MOUSE
|
||||||
| NC score | 0.348141 (rank : 5) | θ value | 0.00228821 (rank : 5) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 218 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01867 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-2B chain C region, membrane-bound form. | |||||
|
IGH1M_MOUSE
|
||||||
| NC score | 0.345139 (rank : 6) | θ value | 0.00228821 (rank : 6) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01869 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig gamma-1 chain C region, membrane-bound form. | |||||
|
MUC_MOUSE
|
||||||
| NC score | 0.327894 (rank : 7) | θ value | θ > 10 (rank : 36) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 242 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01872 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig mu chain C region secreted form. | |||||
|
IGHDM_MOUSE
|
||||||
| NC score | 0.323745 (rank : 8) | θ value | 0.0252991 (rank : 7) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | P01882 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig delta chain C region membrane-bound form. | |||||
|
MUC_HUMAN
|
||||||
| NC score | 0.307039 (rank : 9) | θ value | θ > 10 (rank : 35) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01871 | Gene names | IGHM | |||
|
Domain Architecture |

|
|||||
| Description | Ig mu chain C region. | |||||
|
GCAA_MOUSE
|
||||||
| NC score | 0.302574 (rank : 10) | θ value | θ > 10 (rank : 11) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 210 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01863 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-2A chain C region, A allele. | |||||
|
GCAB_MOUSE
|
||||||
| NC score | 0.300809 (rank : 11) | θ value | θ > 10 (rank : 12) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 203 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01864 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig gamma-2A chain C region secreted form (B allele). | |||||
|
MUCB_HUMAN
|
||||||
| NC score | 0.300663 (rank : 12) | θ value | θ > 10 (rank : 34) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 233 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P04220 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig mu heavy chain disease protein (BOT). | |||||
|
GCB_MOUSE
|
||||||
| NC score | 0.299594 (rank : 13) | θ value | θ > 10 (rank : 13) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01866 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig gamma-2B chain C region secreted form. | |||||
|
IGHG1_MOUSE
|
||||||
| NC score | 0.298803 (rank : 14) | θ value | θ > 10 (rank : 22) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 219 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01868 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-1 chain C region secreted form. | |||||
|
GC3_MOUSE
|
||||||
| NC score | 0.296321 (rank : 15) | θ value | θ > 10 (rank : 10) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 212 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P22436 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig gamma-3 chain C region, secreted form. | |||||
|
IGHE_HUMAN
|
||||||
| NC score | 0.293042 (rank : 16) | θ value | θ > 10 (rank : 19) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 217 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01854 | Gene names | IGHE | |||
|
Domain Architecture |

|
|||||
| Description | Ig epsilon chain C region. | |||||
|
IGHG4_HUMAN
|
||||||
| NC score | 0.287315 (rank : 17) | θ value | θ > 10 (rank : 25) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01861 | Gene names | IGHG4 | |||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-4 chain C region. | |||||
|
IGHG2_HUMAN
|
||||||
| NC score | 0.287288 (rank : 18) | θ value | θ > 10 (rank : 23) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 208 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01859 | Gene names | IGHG2 | |||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-2 chain C region. | |||||
|
IGHG1_HUMAN
|
||||||
| NC score | 0.285387 (rank : 19) | θ value | θ > 10 (rank : 21) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 229 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01857 | Gene names | IGHG1 | |||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-1 chain C region. | |||||
|
IGHG3_HUMAN
|
||||||
| NC score | 0.279376 (rank : 20) | θ value | θ > 10 (rank : 24) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 234 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01860 | Gene names | IGHG3 | |||
|
Domain Architecture |

|
|||||
| Description | Ig gamma-3 chain C region (Heavy chain disease protein) (HDC). | |||||
|
IGHD_HUMAN
|
||||||
| NC score | 0.268298 (rank : 21) | θ value | θ > 10 (rank : 17) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01880 | Gene names | IGHD | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig delta chain C region. | |||||
|
IGHA2_HUMAN
|
||||||
| NC score | 0.267282 (rank : 22) | θ value | θ > 10 (rank : 15) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 184 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01877 | Gene names | IGHA2 | |||
|
Domain Architecture |

|
|||||
| Description | Ig alpha-2 chain C region. | |||||
|
IGHA1_HUMAN
|
||||||
| NC score | 0.266453 (rank : 23) | θ value | θ > 10 (rank : 14) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01876 | Gene names | IGHA1 | |||
|
Domain Architecture |

|
|||||
| Description | Ig alpha-1 chain C region. | |||||
|
IGHE_MOUSE
|
||||||
| NC score | 0.263871 (rank : 24) | θ value | θ > 10 (rank : 20) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 209 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P06336, P01856 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig epsilon chain C region. | |||||
|
IGHA_MOUSE
|
||||||
| NC score | 0.257409 (rank : 25) | θ value | θ > 10 (rank : 16) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01878 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig alpha chain C region. | |||||
|
IGHD_MOUSE
|
||||||
| NC score | 0.191438 (rank : 26) | θ value | θ > 10 (rank : 18) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 160 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01881 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ig delta chain C region secreted form. | |||||
|
KAC_HUMAN
|
||||||
| NC score | 0.173055 (rank : 27) | θ value | θ > 10 (rank : 28) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01834 | Gene names | IGKC | |||
|
Domain Architecture |

|
|||||
| Description | Ig kappa chain C region. | |||||
|
LAC3_MOUSE
|
||||||
| NC score | 0.172649 (rank : 28) | θ value | θ > 10 (rank : 32) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01845 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig lambda-3 chain C region. | |||||
|
KAC_MOUSE
|
||||||
| NC score | 0.164915 (rank : 29) | θ value | θ > 10 (rank : 29) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01837 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig kappa chain C region. | |||||
|
LAC2_MOUSE
|
||||||
| NC score | 0.164878 (rank : 30) | θ value | θ > 10 (rank : 31) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 197 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01844 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig lambda-2 chain C region. | |||||
|
LAC1_MOUSE
|
||||||
| NC score | 0.153085 (rank : 31) | θ value | θ > 10 (rank : 30) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 194 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01843 | Gene names | ||||
|
Domain Architecture |

|
|||||
| Description | Ig lambda-1 chain C region. | |||||
|
LAC_HUMAN
|
||||||
| NC score | 0.152849 (rank : 32) | θ value | θ > 10 (rank : 33) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 195 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01842, P80423 | Gene names | IGLC1 | |||
|
Domain Architecture |

|
|||||
| Description | Ig lambda chain C regions. | |||||
|
IGLL1_MOUSE
|
||||||
| NC score | 0.140157 (rank : 33) | θ value | θ > 10 (rank : 27) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 196 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20764, Q5W1K3 | Gene names | Igll1, Igl-5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Immunoglobulin lambda-like polypeptide 1 precursor (Ig lambda-5) (CD179b antigen). | |||||
|
IGLL1_HUMAN
|
||||||
| NC score | 0.138296 (rank : 34) | θ value | θ > 10 (rank : 26) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 202 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P15814 | Gene names | IGLL1, IGL1 | |||
|
Domain Architecture |

|
|||||
| Description | Immunoglobulin lambda-like polypeptide 1 precursor (Immunoglobulin- related protein 14.1) (Immunoglobulin omega polypeptide) (Ig lambda-5) (CD179b antigen). | |||||
|
TCB_HUMAN
|
||||||
| NC score | 0.134823 (rank : 35) | θ value | θ > 10 (rank : 43) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 172 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01850 | Gene names | TRBC1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell receptor beta chain C region. | |||||
|
TCB2_MOUSE
|
||||||
| NC score | 0.133383 (rank : 36) | θ value | θ > 10 (rank : 42) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01851 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell receptor beta-2 chain C region. | |||||
|
TCB1_MOUSE
|
||||||
| NC score | 0.131171 (rank : 37) | θ value | θ > 10 (rank : 41) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 162 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P01852 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell receptor beta-1 chain C region. | |||||
|
SIRPG_HUMAN
|
||||||
| NC score | 0.102323 (rank : 38) | θ value | θ > 10 (rank : 40) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 359 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9P1W8, Q8WWA5, Q9NQK8 | Gene names | SIRPG, SIRPB2 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-regulatory protein gamma precursor (Signal-regulatory protein beta-2) (SIRP-beta-2) (SIRP-b2) (CD172g antigen). | |||||
|
SHPS1_MOUSE
|
||||||
| NC score | 0.101570 (rank : 39) | θ value | θ > 10 (rank : 38) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 439 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P97797, O08907, O35924, O88555, O88556, P97796, Q8R559, Q9QX57, Q9WTN4 | Gene names | Sirpa, Bit, Myd1, Ptpns1, Shps1, Sirp | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type substrate 1 precursor (SHP substrate 1) (SHPS-1) (Inhibitory receptor SHPS-1) (Signal- regulatory protein alpha-1) (Sirp-alpha-1) (mSIRP-alpha1) (MyD-1 antigen) (Brain Ig-like molecule with tyrosine-based activation motifs) (Bit) (p84) (CD172a antigen). | |||||
|
SIRB1_HUMAN
|
||||||
| NC score | 0.086822 (rank : 40) | θ value | θ > 10 (rank : 39) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 422 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | O00241, Q5TFR0, Q8TB12, Q9H1U5, Q9Y4V0 | Gene names | SIRPB1 | |||
|
Domain Architecture |

|
|||||
| Description | Signal-regulatory protein beta-1 precursor (SIRP-beta-1) (CD172b antigen). | |||||
|
SHPS1_HUMAN
|
||||||
| NC score | 0.080293 (rank : 41) | θ value | θ > 10 (rank : 37) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P78324, O00683, O43799, Q8N517, Q8TAL8, Q9H0Z2, Q9UDX2, Q9UIJ6, Q9Y4U9 | Gene names | SIRPA, BIT, MFR, MYD1, PTPNS1, SHPS1, SIRP | |||
|
Domain Architecture |

|
|||||
| Description | Tyrosine-protein phosphatase non-receptor type substrate 1 precursor (SHP substrate 1) (SHPS-1) (Inhibitory receptor SHPS-1) (Signal- regulatory protein alpha-1) (Sirp-alpha-1) (Sirp-alpha-2) (Sirp-alpha- 3) (MyD-1 antigen) (Brain Ig-like molecule with tyrosine-based activation motifs) (Bit) (Macrophage fusion receptor) (p84) (CD172a antigen). | |||||
|
TCC1_MOUSE
|
||||||
| NC score | 0.072423 (rank : 42) | θ value | θ > 10 (rank : 44) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 56 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P01853 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell receptor gamma chain C region C10.5. | |||||
|
2DMB_MOUSE
|
||||||
| NC score | 0.068926 (rank : 43) | θ value | θ > 10 (rank : 9) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 214 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P35737 | Gene names | H2-DMb1, H-2Mb1, Mb | |||
|
Domain Architecture |

|
|||||
| Description | Class II histocompatibility antigen, M beta 1 chain precursor (H2-M beta 1 chain). | |||||
|
TPSNR_MOUSE
|
||||||
| NC score | 0.063137 (rank : 44) | θ value | θ > 10 (rank : 48) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 336 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8VD31 | Gene names | Tapbpl | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tapasin-related protein precursor (TAPASIN-R) (Tapasin-like) (TAP- binding protein-related protein) (TAPBP-R) (TAP-binding protein-like). | |||||
|
2DMB_HUMAN
|
||||||
| NC score | 0.057143 (rank : 45) | θ value | θ > 10 (rank : 8) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P28068, O77936, Q13012, Q29751, Q9XRX2 | Gene names | HLA-DMB, DMB, RING7 | |||
|
Domain Architecture |

|
|||||
| Description | HLA class II histocompatibility antigen, DM beta chain precursor (MHC class II antigen DMB). | |||||
|
TCC4_MOUSE
|
||||||
| NC score | 0.055703 (rank : 46) | θ value | θ > 10 (rank : 46) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 44 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P06335 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell receptor gamma chain C region 5/10-13. | |||||
|
TCC3_MOUSE
|
||||||
| NC score | 0.051658 (rank : 47) | θ value | θ > 10 (rank : 45) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 49 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P06334 | Gene names | ||||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | T-cell receptor gamma chain C region DFL12. | |||||
|
TPSNR_HUMAN
|
||||||
| NC score | 0.050998 (rank : 48) | θ value | θ > 10 (rank : 47) | |||
| Query Neighborhood Hits | 7 | Target Neighborhood Hits | 330 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q9BX59, Q9NWB8 | Gene names | TAPBPL | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Tapasin-related protein precursor (TAPASIN-R) (Tapasin-like) (TAP- binding protein-related protein) (TAPBP-R) (TAP-binding protein-like). | |||||