Please be patient as the page loads
|
MGST3_MOUSE
|
||||||
| SwissProt Accessions | Q9CPU4, Q3UXC5, Q9D834 | Gene names | Mgst3 | |||
|
Domain Architecture |

|
|||||
| Description | Microsomal glutathione S-transferase 3 (EC 2.5.1.18) (Microsomal GST- 3) (Microsomal GST-III). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
MGST3_MOUSE
|
||||||
| θ value | 1.14559e-87 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CPU4, Q3UXC5, Q9D834 | Gene names | Mgst3 | |||
|
Domain Architecture |

|
|||||
| Description | Microsomal glutathione S-transferase 3 (EC 2.5.1.18) (Microsomal GST- 3) (Microsomal GST-III). | |||||
|
MGST3_HUMAN
|
||||||
| θ value | 2.02684e-76 (rank : 2) | NC score | 0.998178 (rank : 2) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14880, Q6ICN4 | Gene names | MGST3 | |||
|
Domain Architecture |

|
|||||
| Description | Microsomal glutathione S-transferase 3 (EC 2.5.1.18) (Microsomal GST- 3) (Microsomal GST-III). | |||||
|
MGST2_HUMAN
|
||||||
| θ value | 5.62301e-10 (rank : 3) | NC score | 0.474979 (rank : 3) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99735, Q7Z5B8 | Gene names | MGST2, GST2 | |||
|
Domain Architecture |

|
|||||
| Description | Microsomal glutathione S-transferase 2 (EC 2.5.1.18) (Microsomal GST- 2) (Microsomal GST-II). | |||||
|
PTGES_MOUSE
|
||||||
| θ value | 1.06291 (rank : 4) | NC score | 0.103122 (rank : 7) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JM51, Q3T9C5 | Gene names | Ptges, Pges | |||
|
Domain Architecture |
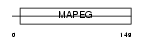
|
|||||
| Description | Prostaglandin E synthase (EC 5.3.99.3) (mPGES-1). | |||||
|
FLAP_MOUSE
|
||||||
| θ value | 4.03905 (rank : 5) | NC score | 0.128782 (rank : 4) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30355, Q9D138 | Gene names | Alox5ap, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arachidonate 5-lipoxygenase-activating protein (FLAP) (MK-886-binding protein). | |||||
|
ADA17_HUMAN
|
||||||
| θ value | 5.27518 (rank : 6) | NC score | 0.023532 (rank : 11) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78536, O60226 | Gene names | ADAM17, CSVP, TACE | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 17 precursor (EC 3.4.24.86) (A disintegrin and metalloproteinase domain 17) (TNF-alpha-converting enzyme) (TNF-alpha convertase) (Snake venom-like protease) (CD156b antigen). | |||||
|
ADA17_MOUSE
|
||||||
| θ value | 5.27518 (rank : 7) | NC score | 0.023776 (rank : 10) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0F8, O88726, Q9R1U4, Q9Z0K3 | Gene names | Adam17, Tace | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 17 precursor (EC 3.4.24.86) (A disintegrin and metalloproteinase domain 17) (TNF-alpha-converting enzyme) (TNF-alpha convertase) (CD156b antigen). | |||||
|
PTGES_HUMAN
|
||||||
| θ value | 6.88961 (rank : 8) | NC score | 0.068854 (rank : 9) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14684, O14900 | Gene names | PTGES, MGST1L1, PGES, PIG12 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin E synthase (EC 5.3.99.3) (Microsomal glutathione S- transferase 1-like 1) (MGST1-L1) (p53-induced apoptosis protein 12). | |||||
|
FLAP_HUMAN
|
||||||
| θ value | 8.99809 (rank : 9) | NC score | 0.121052 (rank : 5) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20292, Q5VV04 | Gene names | ALOX5AP, FLAP | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 5-lipoxygenase-activating protein (FLAP) (MK-886-binding protein). | |||||
|
LTC4S_HUMAN
|
||||||
| θ value | θ > 10 (rank : 10) | NC score | 0.108776 (rank : 6) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16873, Q8N6P0 | Gene names | LTC4S | |||
|
Domain Architecture |

|
|||||
| Description | Leukotriene C4 synthase (EC 4.4.1.20) (Leukotriene-C(4) synthase) (LTC4 synthase). | |||||
|
LTC4S_MOUSE
|
||||||
| θ value | θ > 10 (rank : 11) | NC score | 0.102859 (rank : 8) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60860 | Gene names | Ltc4s | |||
|
Domain Architecture |

|
|||||
| Description | Leukotriene C4 synthase (EC 4.4.1.20) (Leukotriene-C(4) synthase) (LTC4 synthase). | |||||
|
MGST3_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.14559e-87 (rank : 1) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9CPU4, Q3UXC5, Q9D834 | Gene names | Mgst3 | |||
|
Domain Architecture |

|
|||||
| Description | Microsomal glutathione S-transferase 3 (EC 2.5.1.18) (Microsomal GST- 3) (Microsomal GST-III). | |||||
|
MGST3_HUMAN
|
||||||
| NC score | 0.998178 (rank : 2) | θ value | 2.02684e-76 (rank : 2) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | O14880, Q6ICN4 | Gene names | MGST3 | |||
|
Domain Architecture |

|
|||||
| Description | Microsomal glutathione S-transferase 3 (EC 2.5.1.18) (Microsomal GST- 3) (Microsomal GST-III). | |||||
|
MGST2_HUMAN
|
||||||
| NC score | 0.474979 (rank : 3) | θ value | 5.62301e-10 (rank : 3) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q99735, Q7Z5B8 | Gene names | MGST2, GST2 | |||
|
Domain Architecture |

|
|||||
| Description | Microsomal glutathione S-transferase 2 (EC 2.5.1.18) (Microsomal GST- 2) (Microsomal GST-II). | |||||
|
FLAP_MOUSE
|
||||||
| NC score | 0.128782 (rank : 4) | θ value | 4.03905 (rank : 5) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P30355, Q9D138 | Gene names | Alox5ap, Flap | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Arachidonate 5-lipoxygenase-activating protein (FLAP) (MK-886-binding protein). | |||||
|
FLAP_HUMAN
|
||||||
| NC score | 0.121052 (rank : 5) | θ value | 8.99809 (rank : 9) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P20292, Q5VV04 | Gene names | ALOX5AP, FLAP | |||
|
Domain Architecture |

|
|||||
| Description | Arachidonate 5-lipoxygenase-activating protein (FLAP) (MK-886-binding protein). | |||||
|
LTC4S_HUMAN
|
||||||
| NC score | 0.108776 (rank : 6) | θ value | θ > 10 (rank : 10) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q16873, Q8N6P0 | Gene names | LTC4S | |||
|
Domain Architecture |

|
|||||
| Description | Leukotriene C4 synthase (EC 4.4.1.20) (Leukotriene-C(4) synthase) (LTC4 synthase). | |||||
|
PTGES_MOUSE
|
||||||
| NC score | 0.103122 (rank : 7) | θ value | 1.06291 (rank : 4) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9JM51, Q3T9C5 | Gene names | Ptges, Pges | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin E synthase (EC 5.3.99.3) (mPGES-1). | |||||
|
LTC4S_MOUSE
|
||||||
| NC score | 0.102859 (rank : 8) | θ value | θ > 10 (rank : 11) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 5 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q60860 | Gene names | Ltc4s | |||
|
Domain Architecture |

|
|||||
| Description | Leukotriene C4 synthase (EC 4.4.1.20) (Leukotriene-C(4) synthase) (LTC4 synthase). | |||||
|
PTGES_HUMAN
|
||||||
| NC score | 0.068854 (rank : 9) | θ value | 6.88961 (rank : 8) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 6 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O14684, O14900 | Gene names | PTGES, MGST1L1, PGES, PIG12 | |||
|
Domain Architecture |

|
|||||
| Description | Prostaglandin E synthase (EC 5.3.99.3) (Microsomal glutathione S- transferase 1-like 1) (MGST1-L1) (p53-induced apoptosis protein 12). | |||||
|
ADA17_MOUSE
|
||||||
| NC score | 0.023776 (rank : 10) | θ value | 5.27518 (rank : 7) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 201 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0F8, O88726, Q9R1U4, Q9Z0K3 | Gene names | Adam17, Tace | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 17 precursor (EC 3.4.24.86) (A disintegrin and metalloproteinase domain 17) (TNF-alpha-converting enzyme) (TNF-alpha convertase) (CD156b antigen). | |||||
|
ADA17_HUMAN
|
||||||
| NC score | 0.023532 (rank : 11) | θ value | 5.27518 (rank : 6) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 223 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P78536, O60226 | Gene names | ADAM17, CSVP, TACE | |||
|
Domain Architecture |

|
|||||
| Description | ADAM 17 precursor (EC 3.4.24.86) (A disintegrin and metalloproteinase domain 17) (TNF-alpha-converting enzyme) (TNF-alpha convertase) (Snake venom-like protease) (CD156b antigen). | |||||