Please be patient as the page loads
|
LEG8_MOUSE
|
||||||
| SwissProt Accessions | Q9JL15 | Gene names | Lgals8 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-8 (LGALS-8). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
LEG8_MOUSE
|
||||||
| θ value | 4.23373e-183 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JL15 | Gene names | Lgals8 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-8 (LGALS-8). | |||||
|
LEG8_HUMAN
|
||||||
| θ value | 3.61748e-150 (rank : 2) | NC score | 0.998106 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00214, O15215, Q96B92, Q9H584, Q9H585, Q9UEZ6, Q9UP32, Q9UP33, Q9UP34 | Gene names | LGALS8 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-8 (Gal-8) (Prostate carcinoma tumor antigen 1) (PCTA-1) (Po66 carbohydrate-binding protein) (Po66-CBP). | |||||
|
LEG4_MOUSE
|
||||||
| θ value | 6.56568e-51 (rank : 3) | NC score | 0.944801 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K419, O88353, Q91X74 | Gene names | Lgals4 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-4 (Lactose-binding lectin 4). | |||||
|
LEG4_HUMAN
|
||||||
| θ value | 8.57503e-51 (rank : 4) | NC score | 0.949244 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P56470 | Gene names | LGALS4 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-4 (Lactose-binding lectin 4) (L-36 lactose-binding protein) (L36LBP) (Antigen NY-CO-27). | |||||
|
LEG6_MOUSE
|
||||||
| θ value | 1.23823e-49 (rank : 5) | NC score | 0.944990 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O54891, O88352 | Gene names | Lgals6 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-6. | |||||
|
LEG9_MOUSE
|
||||||
| θ value | 1.04822e-48 (rank : 6) | NC score | 0.942646 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08573, O08572 | Gene names | Lgals9 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-9. | |||||
|
LEG9_HUMAN
|
||||||
| θ value | 1.46607e-42 (rank : 7) | NC score | 0.935648 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00182, O14532, O75028, Q9NQ58 | Gene names | LGALS9 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-9 (HOM-HD-21) (Ecalectin). | |||||
|
LEG12_HUMAN
|
||||||
| θ value | 1.32909e-35 (rank : 8) | NC score | 0.898614 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96DT0, Q96DS9, Q96PR9, Q9H258, Q9H259, Q9NZ02 | Gene names | LGALS12, GRIP1 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-12 (Galectin-related inhibitor of proliferation). | |||||
|
LEG12_MOUSE
|
||||||
| θ value | 2.42779e-29 (rank : 9) | NC score | 0.891026 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91VD1, Q3UW90, Q8CCA4, Q8K2L7, Q9JKX2 | Gene names | Lgals12 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-12. | |||||
|
LEG3_HUMAN
|
||||||
| θ value | 1.13037e-18 (rank : 10) | NC score | 0.838808 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17931, Q16005, Q96J47 | Gene names | LGALS3, MAC2 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-31) (Galactoside-binding protein) (GALBP). | |||||
|
LEG3_MOUSE
|
||||||
| θ value | 2.13179e-17 (rank : 11) | NC score | 0.826838 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P16110 | Gene names | Lgals3 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-34 galactoside-binding lectin). | |||||
|
LEG7_MOUSE
|
||||||
| θ value | 1.29331e-14 (rank : 12) | NC score | 0.864714 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O54974 | Gene names | Lgals7 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-7 (Gal-7). | |||||
|
LEG7_HUMAN
|
||||||
| θ value | 8.38298e-14 (rank : 13) | NC score | 0.861858 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P47929 | Gene names | LGALS7, PIG1 | |||
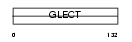
|
Domain Architecture |
|
|||||
| Description | Galectin-7 (Gal-7) (HKL-14) (PI7) (p53-induced protein 1). | |||||
|
LPPL_HUMAN
|
||||||
| θ value | 1.9326e-10 (rank : 14) | NC score | 0.729197 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05315 | Gene names | CLC | |||
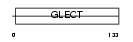
|
Domain Architecture |
|
|||||
| Description | Eosinophil lysophospholipase (EC 3.1.1.5) (Charcot-Leyden crystal protein) (Lysolecithin acylhydrolase) (CLC) (Galectin-10). | |||||
|
LEG1_MOUSE
|
||||||
| θ value | 4.76016e-09 (rank : 15) | NC score | 0.702782 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P16045, P05163, P11946, P17601, Q4FZH4, Q99M27, Q9D0X0 | Gene names | Lgals1, Gbp | |||
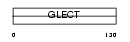
|
Domain Architecture |
|
|||||
| Description | Galectin-1 (Lectin galactoside-binding soluble 1) (Beta-galactoside- binding lectin L-14-I) (Lactose-binding lectin 1) (S-Lac lectin 1) (Galaptin) (14 kDa lectin). | |||||
|
LEG1_HUMAN
|
||||||
| θ value | 8.11959e-09 (rank : 16) | NC score | 0.686940 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09382 | Gene names | LGALS1 | |||
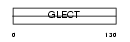
|
Domain Architecture |
|
|||||
| Description | Galectin-1 (Lectin galactoside-binding soluble 1) (Beta-galactoside- binding lectin L-14-I) (Lactose-binding lectin 1) (S-Lac lectin 1) (Galaptin) (14 kDa lectin) (HPL) (HBL) (Putative MAPK-activating protein MP12). | |||||
|
PP13_HUMAN
|
||||||
| θ value | 4.0297e-08 (rank : 17) | NC score | 0.699878 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
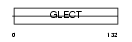
| SwissProt Accessions | Q9UHV8 | Gene names | LGALS13, PLAC8 | |||
|
Domain Architecture |
|
|||||
| Description | Galactoside-binding soluble lectin 13 (EC 3.1.1.5) (Placental tissue protein 13) (Placenta protein 13) (PP13) (Galectin-13). | |||||
|
LEG2_MOUSE
|
||||||
| θ value | 5.26297e-08 (rank : 18) | NC score | 0.700462 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
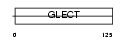
| SwissProt Accessions | Q9CQW5, Q9D800, Q9D862 | Gene names | Lgals2 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-2. | |||||
|
LEG2_HUMAN
|
||||||
| θ value | 1.17247e-07 (rank : 19) | NC score | 0.655838 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
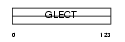
| SwissProt Accessions | P05162 | Gene names | LGALS2 | |||
|
Domain Architecture |
|
|||||
| Description | Galectin-2 (Beta-galactoside-binding lectin L-14-II) (Lactose-binding lectin 2) (S-Lac lectin 2) (HL14). | |||||
|
PPL13_HUMAN
|
||||||
| θ value | 1.99992e-07 (rank : 20) | NC score | 0.697230 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 16 | |
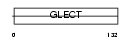
| SwissProt Accessions | Q8TCE9, Q7Z4X8, Q96KD4, Q96KD5, Q96KD6, Q9NR03 | Gene names | LGALS14, PPL13 | |||
|
Domain Architecture |
|
|||||
| Description | Placental protein 13-like (Charcot-Leyden crystal protein 2) (CLC2) (Galectin-14). | |||||
|
LEG8_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 4.23373e-183 (rank : 1) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q9JL15 | Gene names | Lgals8 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-8 (LGALS-8). | |||||
|
LEG8_HUMAN
|
||||||
| NC score | 0.998106 (rank : 2) | θ value | 3.61748e-150 (rank : 2) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 22 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00214, O15215, Q96B92, Q9H584, Q9H585, Q9UEZ6, Q9UP32, Q9UP33, Q9UP34 | Gene names | LGALS8 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-8 (Gal-8) (Prostate carcinoma tumor antigen 1) (PCTA-1) (Po66 carbohydrate-binding protein) (Po66-CBP). | |||||
|
LEG4_HUMAN
|
||||||
| NC score | 0.949244 (rank : 3) | θ value | 8.57503e-51 (rank : 4) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P56470 | Gene names | LGALS4 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-4 (Lactose-binding lectin 4) (L-36 lactose-binding protein) (L36LBP) (Antigen NY-CO-27). | |||||
|
LEG6_MOUSE
|
||||||
| NC score | 0.944990 (rank : 4) | θ value | 1.23823e-49 (rank : 5) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O54891, O88352 | Gene names | Lgals6 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-6. | |||||
|
LEG4_MOUSE
|
||||||
| NC score | 0.944801 (rank : 5) | θ value | 6.56568e-51 (rank : 3) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 32 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q8K419, O88353, Q91X74 | Gene names | Lgals4 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-4 (Lactose-binding lectin 4). | |||||
|
LEG9_MOUSE
|
||||||
| NC score | 0.942646 (rank : 6) | θ value | 1.04822e-48 (rank : 6) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O08573, O08572 | Gene names | Lgals9 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-9. | |||||
|
LEG9_HUMAN
|
||||||
| NC score | 0.935648 (rank : 7) | θ value | 1.46607e-42 (rank : 7) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O00182, O14532, O75028, Q9NQ58 | Gene names | LGALS9 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-9 (HOM-HD-21) (Ecalectin). | |||||
|
LEG12_HUMAN
|
||||||
| NC score | 0.898614 (rank : 8) | θ value | 1.32909e-35 (rank : 8) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q96DT0, Q96DS9, Q96PR9, Q9H258, Q9H259, Q9NZ02 | Gene names | LGALS12, GRIP1 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-12 (Galectin-related inhibitor of proliferation). | |||||
|
LEG12_MOUSE
|
||||||
| NC score | 0.891026 (rank : 9) | θ value | 2.42779e-29 (rank : 9) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q91VD1, Q3UW90, Q8CCA4, Q8K2L7, Q9JKX2 | Gene names | Lgals12 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-12. | |||||
|
LEG7_MOUSE
|
||||||
| NC score | 0.864714 (rank : 10) | θ value | 1.29331e-14 (rank : 12) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 20 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | O54974 | Gene names | Lgals7 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-7 (Gal-7). | |||||
|
LEG7_HUMAN
|
||||||
| NC score | 0.861858 (rank : 11) | θ value | 8.38298e-14 (rank : 13) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P47929 | Gene names | LGALS7, PIG1 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-7 (Gal-7) (HKL-14) (PI7) (p53-induced protein 1). | |||||
|
LEG3_HUMAN
|
||||||
| NC score | 0.838808 (rank : 12) | θ value | 1.13037e-18 (rank : 10) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 124 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P17931, Q16005, Q96J47 | Gene names | LGALS3, MAC2 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-31) (Galactoside-binding protein) (GALBP). | |||||
|
LEG3_MOUSE
|
||||||
| NC score | 0.826838 (rank : 13) | θ value | 2.13179e-17 (rank : 11) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 113 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | P16110 | Gene names | Lgals3 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-3 (Galactose-specific lectin 3) (Mac-2 antigen) (IgE-binding protein) (35 kDa lectin) (Carbohydrate-binding protein 35) (CBP 35) (Laminin-binding protein) (Lectin L-29) (L-34 galactoside-binding lectin). | |||||
|
LPPL_HUMAN
|
||||||
| NC score | 0.729197 (rank : 14) | θ value | 1.9326e-10 (rank : 14) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 21 | Shared Neighborhood Hits | 20 | |
| SwissProt Accessions | Q05315 | Gene names | CLC | |||
|
Domain Architecture |

|
|||||
| Description | Eosinophil lysophospholipase (EC 3.1.1.5) (Charcot-Leyden crystal protein) (Lysolecithin acylhydrolase) (CLC) (Galectin-10). | |||||
|
LEG1_MOUSE
|
||||||
| NC score | 0.702782 (rank : 15) | θ value | 4.76016e-09 (rank : 15) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P16045, P05163, P11946, P17601, Q4FZH4, Q99M27, Q9D0X0 | Gene names | Lgals1, Gbp | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-1 (Lectin galactoside-binding soluble 1) (Beta-galactoside- binding lectin L-14-I) (Lactose-binding lectin 1) (S-Lac lectin 1) (Galaptin) (14 kDa lectin). | |||||
|
LEG2_MOUSE
|
||||||
| NC score | 0.700462 (rank : 16) | θ value | 5.26297e-08 (rank : 18) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9CQW5, Q9D800, Q9D862 | Gene names | Lgals2 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-2. | |||||
|
PP13_HUMAN
|
||||||
| NC score | 0.699878 (rank : 17) | θ value | 4.0297e-08 (rank : 17) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | Q9UHV8 | Gene names | LGALS13, PLAC8 | |||
|
Domain Architecture |

|
|||||
| Description | Galactoside-binding soluble lectin 13 (EC 3.1.1.5) (Placental tissue protein 13) (Placenta protein 13) (PP13) (Galectin-13). | |||||
|
PPL13_HUMAN
|
||||||
| NC score | 0.697230 (rank : 18) | θ value | 1.99992e-07 (rank : 20) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 16 | |
| SwissProt Accessions | Q8TCE9, Q7Z4X8, Q96KD4, Q96KD5, Q96KD6, Q9NR03 | Gene names | LGALS14, PPL13 | |||
|
Domain Architecture |

|
|||||
| Description | Placental protein 13-like (Charcot-Leyden crystal protein 2) (CLC2) (Galectin-14). | |||||
|
LEG1_HUMAN
|
||||||
| NC score | 0.686940 (rank : 19) | θ value | 8.11959e-09 (rank : 16) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 19 | Shared Neighborhood Hits | 19 | |
| SwissProt Accessions | P09382 | Gene names | LGALS1 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-1 (Lectin galactoside-binding soluble 1) (Beta-galactoside- binding lectin L-14-I) (Lactose-binding lectin 1) (S-Lac lectin 1) (Galaptin) (14 kDa lectin) (HPL) (HBL) (Putative MAPK-activating protein MP12). | |||||
|
LEG2_HUMAN
|
||||||
| NC score | 0.655838 (rank : 20) | θ value | 1.17247e-07 (rank : 19) | |||
| Query Neighborhood Hits | 20 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 18 | |
| SwissProt Accessions | P05162 | Gene names | LGALS2 | |||
|
Domain Architecture |

|
|||||
| Description | Galectin-2 (Beta-galactoside-binding lectin L-14-II) (Lactose-binding lectin 2) (S-Lac lectin 2) (HL14). | |||||