Please be patient as the page loads
|
KCMB4_MOUSE
|
||||||
| SwissProt Accessions | Q9JIN6 | Gene names | Kcnmb4 | |||
|
Domain Architecture |
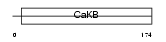
|
|||||
| Description | Calcium-activated potassium channel subunit beta 4 (Calcium-activated potassium channel, subfamily M subunit beta 4) (Maxi K channel subunit beta 4) (BK channel subunit beta 4) (Slo-beta 4) (K(VCA)beta 4) (Charybdotoxin receptor subunit beta 4) (BKbeta4). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
KCMB4_MOUSE
|
||||||
| θ value | 1.8752e-114 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JIN6 | Gene names | Kcnmb4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit beta 4 (Calcium-activated potassium channel, subfamily M subunit beta 4) (Maxi K channel subunit beta 4) (BK channel subunit beta 4) (Slo-beta 4) (K(VCA)beta 4) (Charybdotoxin receptor subunit beta 4) (BKbeta4). | |||||
|
KCMB4_HUMAN
|
||||||
| θ value | 6.24247e-110 (rank : 2) | NC score | 0.999537 (rank : 2) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86W47, Q8IVR3, Q9NPA4, Q9P0G5 | Gene names | KCNMB4 | |||
|
Domain Architecture |
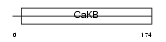
|
|||||
| Description | Calcium-activated potassium channel subunit beta 4 (Calcium-activated potassium channel, subfamily M subunit beta 4) (Maxi K channel subunit beta 4) (BK channel subunit beta 4) (Slo-beta 4) (K(VCA)beta 4) (Charybdotoxin receptor subunit beta 4) (BKbeta4) (Hbeta4). | |||||
|
KCMB3_HUMAN
|
||||||
| θ value | 1.52067e-31 (rank : 3) | NC score | 0.876365 (rank : 3) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NPA1, Q9NPG7, Q9NRM9, Q9UHN3 | Gene names | KCNMB3 | |||
|
Domain Architecture |
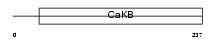
|
|||||
| Description | Calcium-activated potassium channel subunit beta 3 (Calcium-activated potassium channel, subfamily M subunit beta 3) (Maxi K channel subunit beta 3) (BK channel subunit beta 3) (Slo-beta 3) (K(VCA)beta 3) (Charybdotoxin receptor subunit beta 3) (BKbeta3) (Hbeta3). | |||||
|
KCMB2_HUMAN
|
||||||
| θ value | 8.63488e-27 (rank : 4) | NC score | 0.820827 (rank : 5) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y691 | Gene names | KCNMB2 | |||
|
Domain Architecture |
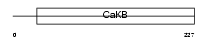
|
|||||
| Description | Calcium-activated potassium channel subunit beta 2 (Calcium-activated potassium channel, subfamily M subunit beta 2) (Maxi K channel subunit beta 2) (BK channel subunit beta 2) (Slo-beta 2) (K(VCA)beta 2) (Charybdotoxin receptor subunit beta 2) (BKbeta2) (Hbeta2) (Hbeta3). | |||||
|
KCMB2_MOUSE
|
||||||
| θ value | 8.63488e-27 (rank : 5) | NC score | 0.821150 (rank : 4) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CZM9 | Gene names | Kcnmb2 | |||
|
Domain Architecture |
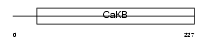
|
|||||
| Description | Calcium-activated potassium channel subunit beta 2 (Calcium-activated potassium channel, subfamily M subunit beta 2) (Maxi K channel subunit beta 2) (BK channel subunit beta 2) (Slo-beta 2) (K(VCA)beta 2) (Charybdotoxin receptor subunit beta 2) (BKbeta2). | |||||
|
KCMB1_HUMAN
|
||||||
| θ value | 8.40245e-06 (rank : 6) | NC score | 0.599391 (rank : 6) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16558, O00707, O00708, P78475, Q8TAX3, Q93005 | Gene names | KCNMB1 | |||
|
Domain Architecture |
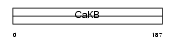
|
|||||
| Description | Calcium-activated potassium channel subunit beta 1 (Calcium-activated potassium channel, subfamily M subunit beta 1) (Maxi K channel subunit beta 1) (BK channel subunit beta 1) (Slo-beta 1) (K(VCA)beta 1) (Charybdotoxin receptor subunit beta 1) (BKbeta1) (Hbeta1) (Calcium- activated potassium channel subunit beta) (BKbeta) (Slo-beta). | |||||
|
KCMB1_MOUSE
|
||||||
| θ value | 5.44631e-05 (rank : 7) | NC score | 0.591828 (rank : 7) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CAE3, O35336, O35645 | Gene names | Kcnmb1 | |||
|
Domain Architecture |
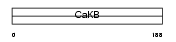
|
|||||
| Description | Calcium-activated potassium channel subunit beta 1 (Calcium-activated potassium channel, subfamily M subunit beta 1) (Maxi K channel subunit beta 1) (BK channel subunit beta 1) (Slo-beta 1) (K(VCA)beta 1) (Charybdotoxin receptor subunit beta 1) (BKbeta1) (Calcium-activated potassium channel subunit beta) (BKbeta) (Slo-beta). | |||||
|
MUC5B_HUMAN
|
||||||
| θ value | 3.0926 (rank : 8) | NC score | 0.011811 (rank : 9) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||
|
C8AP2_HUMAN
|
||||||
| θ value | 6.88961 (rank : 9) | NC score | 0.019034 (rank : 8) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
KCMB4_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 1.8752e-114 (rank : 1) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 9 | |
| SwissProt Accessions | Q9JIN6 | Gene names | Kcnmb4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit beta 4 (Calcium-activated potassium channel, subfamily M subunit beta 4) (Maxi K channel subunit beta 4) (BK channel subunit beta 4) (Slo-beta 4) (K(VCA)beta 4) (Charybdotoxin receptor subunit beta 4) (BKbeta4). | |||||
|
KCMB4_HUMAN
|
||||||
| NC score | 0.999537 (rank : 2) | θ value | 6.24247e-110 (rank : 2) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q86W47, Q8IVR3, Q9NPA4, Q9P0G5 | Gene names | KCNMB4 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit beta 4 (Calcium-activated potassium channel, subfamily M subunit beta 4) (Maxi K channel subunit beta 4) (BK channel subunit beta 4) (Slo-beta 4) (K(VCA)beta 4) (Charybdotoxin receptor subunit beta 4) (BKbeta4) (Hbeta4). | |||||
|
KCMB3_HUMAN
|
||||||
| NC score | 0.876365 (rank : 3) | θ value | 1.52067e-31 (rank : 3) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9NPA1, Q9NPG7, Q9NRM9, Q9UHN3 | Gene names | KCNMB3 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit beta 3 (Calcium-activated potassium channel, subfamily M subunit beta 3) (Maxi K channel subunit beta 3) (BK channel subunit beta 3) (Slo-beta 3) (K(VCA)beta 3) (Charybdotoxin receptor subunit beta 3) (BKbeta3) (Hbeta3). | |||||
|
KCMB2_MOUSE
|
||||||
| NC score | 0.821150 (rank : 4) | θ value | 8.63488e-27 (rank : 5) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9CZM9 | Gene names | Kcnmb2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit beta 2 (Calcium-activated potassium channel, subfamily M subunit beta 2) (Maxi K channel subunit beta 2) (BK channel subunit beta 2) (Slo-beta 2) (K(VCA)beta 2) (Charybdotoxin receptor subunit beta 2) (BKbeta2). | |||||
|
KCMB2_HUMAN
|
||||||
| NC score | 0.820827 (rank : 5) | θ value | 8.63488e-27 (rank : 4) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q9Y691 | Gene names | KCNMB2 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit beta 2 (Calcium-activated potassium channel, subfamily M subunit beta 2) (Maxi K channel subunit beta 2) (BK channel subunit beta 2) (Slo-beta 2) (K(VCA)beta 2) (Charybdotoxin receptor subunit beta 2) (BKbeta2) (Hbeta2) (Hbeta3). | |||||
|
KCMB1_HUMAN
|
||||||
| NC score | 0.599391 (rank : 6) | θ value | 8.40245e-06 (rank : 6) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q16558, O00707, O00708, P78475, Q8TAX3, Q93005 | Gene names | KCNMB1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit beta 1 (Calcium-activated potassium channel, subfamily M subunit beta 1) (Maxi K channel subunit beta 1) (BK channel subunit beta 1) (Slo-beta 1) (K(VCA)beta 1) (Charybdotoxin receptor subunit beta 1) (BKbeta1) (Hbeta1) (Calcium- activated potassium channel subunit beta) (BKbeta) (Slo-beta). | |||||
|
KCMB1_MOUSE
|
||||||
| NC score | 0.591828 (rank : 7) | θ value | 5.44631e-05 (rank : 7) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 7 | Shared Neighborhood Hits | 7 | |
| SwissProt Accessions | Q8CAE3, O35336, O35645 | Gene names | Kcnmb1 | |||
|
Domain Architecture |

|
|||||
| Description | Calcium-activated potassium channel subunit beta 1 (Calcium-activated potassium channel, subfamily M subunit beta 1) (Maxi K channel subunit beta 1) (BK channel subunit beta 1) (Slo-beta 1) (K(VCA)beta 1) (Charybdotoxin receptor subunit beta 1) (BKbeta1) (Calcium-activated potassium channel subunit beta) (BKbeta) (Slo-beta). | |||||
|
C8AP2_HUMAN
|
||||||
| NC score | 0.019034 (rank : 8) | θ value | 6.88961 (rank : 9) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 548 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9UKL3, Q6PH76, Q7LCQ7, Q86YD9, Q9NUQ4, Q9NZV9, Q9P2N1, Q9Y563 | Gene names | CASP8AP2, FLASH, KIAA1315, RIP25 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | CASP8-associated protein 2 (FLICE-associated huge protein). | |||||
|
MUC5B_HUMAN
|
||||||
| NC score | 0.011811 (rank : 9) | θ value | 3.0926 (rank : 8) | |||
| Query Neighborhood Hits | 9 | Target Neighborhood Hits | 1020 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9HC84, O00447, O00573, O14985, O15494, O95291, O95451, Q14881, Q99552, Q9UE28 | Gene names | MUC5B, MUC5 | |||
|
Domain Architecture |

|
|||||
| Description | Mucin-5B precursor (Mucin-5 subtype B, tracheobronchial) (High molecular weight salivary mucin MG1) (Sublingual gland mucin). | |||||