Please be patient as the page loads
|
K0859_MOUSE
|
||||||
| SwissProt Accessions | Q91YR5, Q3TFY3, Q69ZX4, Q8BWN2, Q8BX81 | Gene names | Kiaa0859 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0859. | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
K0859_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.996384 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N6R0, O94940, Q5TGP9, Q5TGQ0, Q8N2P8, Q96J11, Q96SQ0, Q9Y2Z1, Q9Y3M6 | Gene names | KIAA0859 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0859. | |||||
|
K0859_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91YR5, Q3TFY3, Q69ZX4, Q8BWN2, Q8BX81 | Gene names | Kiaa0859 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0859. | |||||
|
ECE2_HUMAN
|
||||||
| θ value | 4.92598e-06 (rank : 3) | NC score | 0.121849 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60344, Q96NX3, Q96NX4 | Gene names | ECE2, KIAA0604 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme 2 (EC 3.4.24.71) (ECE-2). | |||||
|
GOGA4_HUMAN
|
||||||
| θ value | 0.0330416 (rank : 4) | NC score | 0.019710 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
COQ5_HUMAN
|
||||||
| θ value | 0.125558 (rank : 5) | NC score | 0.121285 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5HYK3, Q32Q28, Q53HH0, Q96LV1, Q9BSP8 | Gene names | COQ5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquinone biosynthesis methyltransferase COQ5, mitochondrial precursor (EC 2.1.1.-). | |||||
|
COQ5_MOUSE
|
||||||
| θ value | 1.06291 (rank : 6) | NC score | 0.102027 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CXI0, Q9D6Y6, Q9D900 | Gene names | Coq5, D5Ertd33e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquinone biosynthesis methyltransferase COQ5, mitochondrial precursor (EC 2.1.1.-). | |||||
|
MYCB2_MOUSE
|
||||||
| θ value | 2.36792 (rank : 7) | NC score | 0.021622 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
NED4L_HUMAN
|
||||||
| θ value | 3.0926 (rank : 8) | NC score | 0.009382 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
GEMI_MOUSE
|
||||||
| θ value | 4.03905 (rank : 9) | NC score | 0.030968 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88513 | Gene names | Gmnn | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
TMPS2_HUMAN
|
||||||
| θ value | 5.27518 (rank : 10) | NC score | 0.004217 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15393, Q9BXX1 | Gene names | TMPRSS2, PRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 precursor (EC 3.4.21.-) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
ZN165_HUMAN
|
||||||
| θ value | 5.27518 (rank : 11) | NC score | 0.000863 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 734 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49910 | Gene names | ZNF165, ZPF165, ZSCAN7 | |||
|
Domain Architecture |
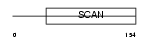
|
|||||
| Description | Zinc finger protein 165 (LD65) (Zinc finger and SCAN domain-containing protein 7). | |||||
|
FYCO1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 12) | NC score | 0.008834 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
MACF4_HUMAN
|
||||||
| θ value | 6.88961 (rank : 13) | NC score | 0.009587 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
PDCD8_HUMAN
|
||||||
| θ value | 6.88961 (rank : 14) | NC score | 0.027092 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95831, Q6I9X6, Q9Y3I3, Q9Y3I4 | Gene names | PDCD8, AIF | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 8, mitochondrial precursor (EC 1.-.-.-) (Apoptosis-inducing factor). | |||||
|
PDCD8_MOUSE
|
||||||
| θ value | 6.88961 (rank : 15) | NC score | 0.027089 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0X1 | Gene names | Pdcd8, Aif | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 8, mitochondrial precursor (EC 1.-.-.-) (Apoptosis-inducing factor). | |||||
|
ARHGG_MOUSE
|
||||||
| θ value | 8.99809 (rank : 16) | NC score | 0.007623 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U5C8, Q501M8, Q8VCE8 | Gene names | Arhgef16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
COQ3_HUMAN
|
||||||
| θ value | 8.99809 (rank : 17) | NC score | 0.029098 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZJ6, Q6P4F0, Q8IXG6, Q96BG1, Q9H0N1 | Gene names | COQ3 | |||
|
Domain Architecture |
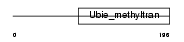
|
|||||
| Description | Hexaprenyldihydroxybenzoate methyltransferase, mitochondrial precursor (EC 2.1.1.114) (Dihydroxyhexaprenylbenzoate methyltransferase) (3,4- dihydroxy-5-hexaprenylbenzoate methyltransferase) (DHHB methyltransferase) (DHHB-MT) (DHHB-MTase). | |||||
|
K0859_MOUSE
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 17 | |
| SwissProt Accessions | Q91YR5, Q3TFY3, Q69ZX4, Q8BWN2, Q8BX81 | Gene names | Kiaa0859 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0859. | |||||
|
K0859_HUMAN
|
||||||
| NC score | 0.996384 (rank : 2) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | Q8N6R0, O94940, Q5TGP9, Q5TGQ0, Q8N2P8, Q96J11, Q96SQ0, Q9Y2Z1, Q9Y3M6 | Gene names | KIAA0859 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein KIAA0859. | |||||
|
ECE2_HUMAN
|
||||||
| NC score | 0.121849 (rank : 3) | θ value | 4.92598e-06 (rank : 3) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O60344, Q96NX3, Q96NX4 | Gene names | ECE2, KIAA0604 | |||
|
Domain Architecture |

|
|||||
| Description | Endothelin-converting enzyme 2 (EC 3.4.24.71) (ECE-2). | |||||
|
COQ5_HUMAN
|
||||||
| NC score | 0.121285 (rank : 4) | θ value | 0.125558 (rank : 5) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q5HYK3, Q32Q28, Q53HH0, Q96LV1, Q9BSP8 | Gene names | COQ5 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquinone biosynthesis methyltransferase COQ5, mitochondrial precursor (EC 2.1.1.-). | |||||
|
COQ5_MOUSE
|
||||||
| NC score | 0.102027 (rank : 5) | θ value | 1.06291 (rank : 6) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q9CXI0, Q9D6Y6, Q9D900 | Gene names | Coq5, D5Ertd33e | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Ubiquinone biosynthesis methyltransferase COQ5, mitochondrial precursor (EC 2.1.1.-). | |||||
|
GEMI_MOUSE
|
||||||
| NC score | 0.030968 (rank : 6) | θ value | 4.03905 (rank : 9) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 163 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | O88513 | Gene names | Gmnn | |||
|
Domain Architecture |

|
|||||
| Description | Geminin. | |||||
|
COQ3_HUMAN
|
||||||
| NC score | 0.029098 (rank : 7) | θ value | 8.99809 (rank : 17) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 18 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9NZJ6, Q6P4F0, Q8IXG6, Q96BG1, Q9H0N1 | Gene names | COQ3 | |||
|
Domain Architecture |

|
|||||
| Description | Hexaprenyldihydroxybenzoate methyltransferase, mitochondrial precursor (EC 2.1.1.114) (Dihydroxyhexaprenylbenzoate methyltransferase) (3,4- dihydroxy-5-hexaprenylbenzoate methyltransferase) (DHHB methyltransferase) (DHHB-MT) (DHHB-MTase). | |||||
|
PDCD8_HUMAN
|
||||||
| NC score | 0.027092 (rank : 8) | θ value | 6.88961 (rank : 14) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 13 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O95831, Q6I9X6, Q9Y3I3, Q9Y3I4 | Gene names | PDCD8, AIF | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 8, mitochondrial precursor (EC 1.-.-.-) (Apoptosis-inducing factor). | |||||
|
PDCD8_MOUSE
|
||||||
| NC score | 0.027089 (rank : 9) | θ value | 6.88961 (rank : 15) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9Z0X1 | Gene names | Pdcd8, Aif | |||
|
Domain Architecture |

|
|||||
| Description | Programmed cell death protein 8, mitochondrial precursor (EC 1.-.-.-) (Apoptosis-inducing factor). | |||||
|
MYCB2_MOUSE
|
||||||
| NC score | 0.021622 (rank : 10) | θ value | 2.36792 (rank : 7) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 307 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q7TPH6, Q6PCM8 | Gene names | Mycbp2, Pam, Phr1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Probable ubiquitin ligase protein MYCBP2 (EC 6.3.2.-) (Myc-binding protein 2) (Protein associated with Myc) (Pam/highwire/rpm-1 protein). | |||||
|
GOGA4_HUMAN
|
||||||
| NC score | 0.019710 (rank : 11) | θ value | 0.0330416 (rank : 4) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 2034 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q13439, Q13270, Q13654, Q14436 | Gene names | GOLGA4 | |||
|
Domain Architecture |

|
|||||
| Description | Golgin subfamily A member 4 (Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (Protein 72.1). | |||||
|
MACF4_HUMAN
|
||||||
| NC score | 0.009587 (rank : 12) | θ value | 6.88961 (rank : 13) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1321 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q96PK2, Q8WXY1 | Gene names | MACF1, ABP620, ACF7, KIAA0465, KIAA1251 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-actin cross-linking factor 1, isoform 4. | |||||
|
NED4L_HUMAN
|
||||||
| NC score | 0.009382 (rank : 13) | θ value | 3.0926 (rank : 8) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 238 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q96PU5, O43165, Q7Z5F1, Q7Z5F2, Q7Z5N3, Q8N5A7, Q8WUU9, Q9BW58, Q9H2W4, Q9NT88 | Gene names | NEDD4L, KIAA0439, NEDL3 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | E3 ubiquitin-protein ligase NEDD4-like protein (EC 6.3.2.-) (Nedd4-2) (NEDD4.2). | |||||
|
FYCO1_HUMAN
|
||||||
| NC score | 0.008834 (rank : 14) | θ value | 6.88961 (rank : 12) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 1149 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q9BQS8, Q3MJE6, Q86T41, Q86TB1, Q8TEF9, Q96IV5, Q9H8P9 | Gene names | FYCO1, ZFYVE7 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | FYVE and coiled-coil domain-containing protein 1 (Zinc finger FYVE domain-containing protein 7). | |||||
|
ARHGG_MOUSE
|
||||||
| NC score | 0.007623 (rank : 15) | θ value | 8.99809 (rank : 16) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 152 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q3U5C8, Q501M8, Q8VCE8 | Gene names | Arhgef16 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Rho guanine nucleotide exchange factor 16. | |||||
|
TMPS2_HUMAN
|
||||||
| NC score | 0.004217 (rank : 16) | θ value | 5.27518 (rank : 10) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 252 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | O15393, Q9BXX1 | Gene names | TMPRSS2, PRSS10 | |||
|
Domain Architecture |

|
|||||
| Description | Transmembrane protease, serine 2 precursor (EC 3.4.21.-) [Contains: Transmembrane protease, serine 2 non-catalytic chain; Transmembrane protease, serine 2 catalytic chain]. | |||||
|
ZN165_HUMAN
|
||||||
| NC score | 0.000863 (rank : 17) | θ value | 5.27518 (rank : 11) | |||
| Query Neighborhood Hits | 17 | Target Neighborhood Hits | 734 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P49910 | Gene names | ZNF165, ZPF165, ZSCAN7 | |||
|
Domain Architecture |

|
|||||
| Description | Zinc finger protein 165 (LD65) (Zinc finger and SCAN domain-containing protein 7). | |||||