Please be patient as the page loads
|
IMDH1_HUMAN
|
||||||
| SwissProt Accessions | P20839, Q8N194, Q96NU2 | Gene names | IMPDH1, IMPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Inosine-5'-monophosphate dehydrogenase 1 (EC 1.1.1.205) (IMP dehydrogenase 1) (IMPDH-I) (IMPD 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
IMDH1_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P20839, Q8N194, Q96NU2 | Gene names | IMPDH1, IMPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Inosine-5'-monophosphate dehydrogenase 1 (EC 1.1.1.205) (IMP dehydrogenase 1) (IMPDH-I) (IMPD 1). | |||||
|
IMDH1_MOUSE
|
||||||
| θ value | 0 (rank : 2) | NC score | 0.999784 (rank : 2) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P50096 | Gene names | Impdh1 | |||
|
Domain Architecture |

|
|||||
| Description | Inosine-5'-monophosphate dehydrogenase 1 (EC 1.1.1.205) (IMP dehydrogenase 1) (IMPDH-I) (IMPD 1). | |||||
|
IMDH2_HUMAN
|
||||||
| θ value | 0 (rank : 3) | NC score | 0.998498 (rank : 3) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P12268, Q6LEF3 | Gene names | IMPDH2, IMPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Inosine-5'-monophosphate dehydrogenase 2 (EC 1.1.1.205) (IMP dehydrogenase 2) (IMPDH-II) (IMPD 2). | |||||
|
IMDH2_MOUSE
|
||||||
| θ value | 0 (rank : 4) | NC score | 0.998443 (rank : 4) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24547, Q61734, Q91Z11 | Gene names | Impdh2, Impdh | |||
|
Domain Architecture |

|
|||||
| Description | Inosine-5'-monophosphate dehydrogenase 2 (EC 1.1.1.205) (IMP dehydrogenase 2) (IMPDH-II) (IMPD 2). | |||||
|
GMPR2_HUMAN
|
||||||
| θ value | 7.80994e-28 (rank : 5) | NC score | 0.744012 (rank : 7) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2T1, Q6IAJ8 | Gene names | GMPR2 | |||
|
Domain Architecture |
|
|||||
| Description | GMP reductase 2 (EC 1.7.1.7) (Guanosine 5'-monophosphate oxidoreductase 2) (Guanosine monophosphate reductase 2). | |||||
|
GMPR2_MOUSE
|
||||||
| θ value | 1.33217e-27 (rank : 6) | NC score | 0.742614 (rank : 8) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99L27, Q8R1T5 | Gene names | Gmpr2 | |||
|
Domain Architecture |
|
|||||
| Description | GMP reductase 2 (EC 1.7.1.7) (Guanosine 5'-monophosphate oxidoreductase 2) (Guanosine monophosphate reductase 2). | |||||
|
GMPR1_MOUSE
|
||||||
| θ value | 2.96777e-27 (rank : 7) | NC score | 0.744358 (rank : 5) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DCZ1 | Gene names | Gmpr, Gmpr1 | |||
|
Domain Architecture |
|
|||||
| Description | GMP reductase 1 (EC 1.7.1.7) (Guanosine 5'-monophosphate oxidoreductase 1) (Guanosine monophosphate reductase 1). | |||||
|
GMPR1_HUMAN
|
||||||
| θ value | 3.87602e-27 (rank : 8) | NC score | 0.744348 (rank : 6) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36959, Q96HQ6 | Gene names | GMPR, GMPR1 | |||
|
Domain Architecture |
|
|||||
| Description | GMP reductase 1 (EC 1.7.1.7) (Guanosine 5'-monophosphate oxidoreductase 1) (Guanosine monophosphate reductase 1). | |||||
|
IF2M_HUMAN
|
||||||
| θ value | 0.365318 (rank : 9) | NC score | 0.049515 (rank : 13) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46199 | Gene names | MTIF2 | |||
|
Domain Architecture |
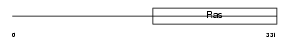
|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
HAOX2_HUMAN
|
||||||
| θ value | 0.813845 (rank : 10) | NC score | 0.080617 (rank : 10) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYQ3, Q9UJS6 | Gene names | HAO2, HAOX2 | |||
|
Domain Architecture |
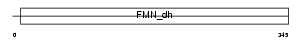
|
|||||
| Description | Hydroxyacid oxidase 2 (EC 1.1.3.15) (HAOX2) ((S)-2-hydroxy-acid oxidase, peroxisomal) (Long chain alpha-hydroxy acid oxidase) (Long- chain L-2-hydroxy acid oxidase). | |||||
|
HAOX2_MOUSE
|
||||||
| θ value | 0.813845 (rank : 11) | NC score | 0.079826 (rank : 11) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYQ2, Q9JHS7, Q9JI00 | Gene names | Hao2, Hao3, Haox2 | |||
|
Domain Architecture |
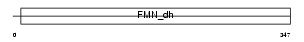
|
|||||
| Description | Hydroxyacid oxidase 2 (EC 1.1.3.15) (HAOX2) ((S)-2-hydroxy-acid oxidase, peroxisomal) (Medium chain alpha-hydroxy acid oxidase) (Medium-chain L-2-hydroxy acid oxidase). | |||||
|
HAOX1_MOUSE
|
||||||
| θ value | 1.38821 (rank : 12) | NC score | 0.085587 (rank : 9) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WU19 | Gene names | Hao1, Gox1, Hao-1 | |||
|
Domain Architecture |
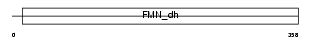
|
|||||
| Description | Hydroxyacid oxidase 1 (EC 1.1.3.15) (HAOX1) (Glycolate oxidase) (GOX). | |||||
|
HAOX1_HUMAN
|
||||||
| θ value | 3.0926 (rank : 13) | NC score | 0.074941 (rank : 12) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UJM8, Q9UPZ0, Q9Y3I7 | Gene names | HAO1, GOX1, HAOX1 | |||
|
Domain Architecture |
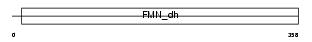
|
|||||
| Description | Hydroxyacid oxidase 1 (EC 1.1.3.15) (HAOX1) (Glycolate oxidase) (GOX). | |||||
|
IF2M_MOUSE
|
||||||
| θ value | 3.0926 (rank : 14) | NC score | 0.036312 (rank : 14) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91YJ5 | Gene names | Mtif2 | |||
|
Domain Architecture |
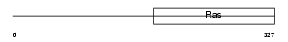
|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
IMDH1_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P20839, Q8N194, Q96NU2 | Gene names | IMPDH1, IMPD1 | |||
|
Domain Architecture |

|
|||||
| Description | Inosine-5'-monophosphate dehydrogenase 1 (EC 1.1.1.205) (IMP dehydrogenase 1) (IMPDH-I) (IMPD 1). | |||||
|
IMDH1_MOUSE
|
||||||
| NC score | 0.999784 (rank : 2) | θ value | 0 (rank : 2) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 16 | Shared Neighborhood Hits | 14 | |
| SwissProt Accessions | P50096 | Gene names | Impdh1 | |||
|
Domain Architecture |

|
|||||
| Description | Inosine-5'-monophosphate dehydrogenase 1 (EC 1.1.1.205) (IMP dehydrogenase 1) (IMPDH-I) (IMPD 1). | |||||
|
IMDH2_HUMAN
|
||||||
| NC score | 0.998498 (rank : 3) | θ value | 0 (rank : 3) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P12268, Q6LEF3 | Gene names | IMPDH2, IMPD2 | |||
|
Domain Architecture |

|
|||||
| Description | Inosine-5'-monophosphate dehydrogenase 2 (EC 1.1.1.205) (IMP dehydrogenase 2) (IMPDH-II) (IMPD 2). | |||||
|
IMDH2_MOUSE
|
||||||
| NC score | 0.998443 (rank : 4) | θ value | 0 (rank : 4) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 15 | Shared Neighborhood Hits | 12 | |
| SwissProt Accessions | P24547, Q61734, Q91Z11 | Gene names | Impdh2, Impdh | |||
|
Domain Architecture |

|
|||||
| Description | Inosine-5'-monophosphate dehydrogenase 2 (EC 1.1.1.205) (IMP dehydrogenase 2) (IMPDH-II) (IMPD 2). | |||||
|
GMPR1_MOUSE
|
||||||
| NC score | 0.744358 (rank : 5) | θ value | 2.96777e-27 (rank : 7) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 10 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9DCZ1 | Gene names | Gmpr, Gmpr1 | |||
|
Domain Architecture |

|
|||||
| Description | GMP reductase 1 (EC 1.7.1.7) (Guanosine 5'-monophosphate oxidoreductase 1) (Guanosine monophosphate reductase 1). | |||||
|
GMPR1_HUMAN
|
||||||
| NC score | 0.744348 (rank : 6) | θ value | 3.87602e-27 (rank : 8) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 12 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | P36959, Q96HQ6 | Gene names | GMPR, GMPR1 | |||
|
Domain Architecture |

|
|||||
| Description | GMP reductase 1 (EC 1.7.1.7) (Guanosine 5'-monophosphate oxidoreductase 1) (Guanosine monophosphate reductase 1). | |||||
|
GMPR2_HUMAN
|
||||||
| NC score | 0.744012 (rank : 7) | θ value | 7.80994e-28 (rank : 5) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9P2T1, Q6IAJ8 | Gene names | GMPR2 | |||
|
Domain Architecture |

|
|||||
| Description | GMP reductase 2 (EC 1.7.1.7) (Guanosine 5'-monophosphate oxidoreductase 2) (Guanosine monophosphate reductase 2). | |||||
|
GMPR2_MOUSE
|
||||||
| NC score | 0.742614 (rank : 8) | θ value | 1.33217e-27 (rank : 6) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q99L27, Q8R1T5 | Gene names | Gmpr2 | |||
|
Domain Architecture |

|
|||||
| Description | GMP reductase 2 (EC 1.7.1.7) (Guanosine 5'-monophosphate oxidoreductase 2) (Guanosine monophosphate reductase 2). | |||||
|
HAOX1_MOUSE
|
||||||
| NC score | 0.085587 (rank : 9) | θ value | 1.38821 (rank : 12) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 9 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9WU19 | Gene names | Hao1, Gox1, Hao-1 | |||
|
Domain Architecture |

|
|||||
| Description | Hydroxyacid oxidase 1 (EC 1.1.3.15) (HAOX1) (Glycolate oxidase) (GOX). | |||||
|
HAOX2_HUMAN
|
||||||
| NC score | 0.080617 (rank : 10) | θ value | 0.813845 (rank : 10) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 11 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYQ3, Q9UJS6 | Gene names | HAO2, HAOX2 | |||
|
Domain Architecture |

|
|||||
| Description | Hydroxyacid oxidase 2 (EC 1.1.3.15) (HAOX2) ((S)-2-hydroxy-acid oxidase, peroxisomal) (Long chain alpha-hydroxy acid oxidase) (Long- chain L-2-hydroxy acid oxidase). | |||||
|
HAOX2_MOUSE
|
||||||
| NC score | 0.079826 (rank : 11) | θ value | 0.813845 (rank : 11) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 14 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9NYQ2, Q9JHS7, Q9JI00 | Gene names | Hao2, Hao3, Haox2 | |||
|
Domain Architecture |

|
|||||
| Description | Hydroxyacid oxidase 2 (EC 1.1.3.15) (HAOX2) ((S)-2-hydroxy-acid oxidase, peroxisomal) (Medium chain alpha-hydroxy acid oxidase) (Medium-chain L-2-hydroxy acid oxidase). | |||||
|
HAOX1_HUMAN
|
||||||
| NC score | 0.074941 (rank : 12) | θ value | 3.0926 (rank : 13) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 8 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q9UJM8, Q9UPZ0, Q9Y3I7 | Gene names | HAO1, GOX1, HAOX1 | |||
|
Domain Architecture |

|
|||||
| Description | Hydroxyacid oxidase 1 (EC 1.1.3.15) (HAOX1) (Glycolate oxidase) (GOX). | |||||
|
IF2M_HUMAN
|
||||||
| NC score | 0.049515 (rank : 13) | θ value | 0.365318 (rank : 9) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 78 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | P46199 | Gene names | MTIF2 | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||
|
IF2M_MOUSE
|
||||||
| NC score | 0.036312 (rank : 14) | θ value | 3.0926 (rank : 14) | |||
| Query Neighborhood Hits | 14 | Target Neighborhood Hits | 183 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q91YJ5 | Gene names | Mtif2 | |||
|
Domain Architecture |

|
|||||
| Description | Translation initiation factor IF-2, mitochondrial precursor (IF-2Mt) (IF-2(Mt)). | |||||