Please be patient as the page loads
|
FA54A_HUMAN
|
||||||
| SwissProt Accessions | Q6P444, Q5JWR7, Q6ZUE8, Q7L3U6, Q9BZ39 | Gene names | FAM54A, DUFD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM54A (DUF729 domain-containing protein 1). | |||||
| Rank Plots |
Jump to hits sorted by NC score
|
|||||
|
FA54A_HUMAN
|
||||||
| θ value | 0 (rank : 1) | NC score | 0.999962 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6P444, Q5JWR7, Q6ZUE8, Q7L3U6, Q9BZ39 | Gene names | FAM54A, DUFD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM54A (DUF729 domain-containing protein 1). | |||||
|
FA54A_MOUSE
|
||||||
| θ value | 1.17728e-108 (rank : 2) | NC score | 0.971031 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VED8 | Gene names | Fam54a, Dufd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM54A (DUF729 domain-containing protein 1). | |||||
|
MTFR1_MOUSE
|
||||||
| θ value | 2.12685e-25 (rank : 3) | NC score | 0.761785 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99MB2, Q9CQZ4 | Gene names | Mtfr1, Kiaa0009 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial fission regulator 1. | |||||
|
MTFR1_HUMAN
|
||||||
| θ value | 3.07116e-24 (rank : 4) | NC score | 0.747268 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15390, Q6IB94, Q86XH5, Q8IVD7 | Gene names | MTFR1, CHPPR, KIAA0009 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial fission regulator 1 (Chondrocyte protein with a poly- proline region). | |||||
|
FA47B_HUMAN
|
||||||
| θ value | 0.00869519 (rank : 5) | NC score | 0.070142 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
PRR11_MOUSE
|
||||||
| θ value | 0.813845 (rank : 6) | NC score | 0.082455 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHE0, Q3V3H1 | Gene names | Prr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 11. | |||||
|
CEBPE_HUMAN
|
||||||
| θ value | 1.81305 (rank : 7) | NC score | 0.026525 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
SFRS4_HUMAN
|
||||||
| θ value | 1.81305 (rank : 8) | NC score | 0.024205 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
SFRS4_MOUSE
|
||||||
| θ value | 1.81305 (rank : 9) | NC score | 0.024473 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
2DMB_HUMAN
|
||||||
| θ value | 2.36792 (rank : 10) | NC score | 0.012187 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28068, O77936, Q13012, Q29751, Q9XRX2 | Gene names | HLA-DMB, DMB, RING7 | |||
|
Domain Architecture |
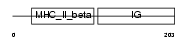
|
|||||
| Description | HLA class II histocompatibility antigen, DM beta chain precursor (MHC class II antigen DMB). | |||||
|
EP15_HUMAN
|
||||||
| θ value | 2.36792 (rank : 11) | NC score | 0.012260 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
CAC1G_HUMAN
|
||||||
| θ value | 3.0926 (rank : 12) | NC score | 0.012107 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
GON4L_MOUSE
|
||||||
| θ value | 4.03905 (rank : 13) | NC score | 0.018657 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
PRR11_HUMAN
|
||||||
| θ value | 5.27518 (rank : 14) | NC score | 0.061722 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96HE9, Q9NUZ7, Q9NXE9 | Gene names | PRR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 11. | |||||
|
RNF12_MOUSE
|
||||||
| θ value | 5.27518 (rank : 15) | NC score | 0.015674 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTV7 | Gene names | Rnf12, Rlim | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM). | |||||
|
TLE3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 16) | NC score | 0.012998 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04726, Q8IVV6, Q8WVR2, Q9HCM5 | Gene names | TLE3, KIAA1547 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG3). | |||||
|
TLE3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 17) | NC score | 0.013092 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08122, Q923A4 | Gene names | Tle3, Esg | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG) (Grg-3). | |||||
|
TSH3_HUMAN
|
||||||
| θ value | 5.27518 (rank : 18) | NC score | 0.031638 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q63HK5, Q9H0G6, Q9P254 | Gene names | TSHZ3, KIAA1474, TSH3, ZNF537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
TSH3_MOUSE
|
||||||
| θ value | 5.27518 (rank : 19) | NC score | 0.031719 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CGV9, Q5DTX4 | Gene names | Tshz3, Kiaa1474, Tsh3, Zfp537, Znf537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
ABCAD_MOUSE
|
||||||
| θ value | 6.88961 (rank : 20) | NC score | 0.007483 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SSE9, Q80T20, Q8BHZ2, Q8CB91 | Gene names | Abca13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
SPRL1_HUMAN
|
||||||
| θ value | 6.88961 (rank : 21) | NC score | 0.014056 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
CLC4F_MOUSE
|
||||||
| θ value | 8.99809 (rank : 22) | NC score | 0.005402 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70194, Q8BLZ8 | Gene names | Clec4f, Clecsf13, Kclr | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13) (Kupffer cell receptor). | |||||
|
MAP2_MOUSE
|
||||||
| θ value | 8.99809 (rank : 23) | NC score | 0.012277 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
TAB2_HUMAN
|
||||||
| θ value | 8.99809 (rank : 24) | NC score | 0.012271 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYJ8, O94838, Q6I9W8, Q76N06, Q9UFP7 | Gene names | MAP3K7IP2, KIAA0733, TAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2). | |||||
|
FA54A_HUMAN
|
||||||
| NC score | 0.999962 (rank : 1) | θ value | 0 (rank : 1) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 24 | Shared Neighborhood Hits | 24 | |
| SwissProt Accessions | Q6P444, Q5JWR7, Q6ZUE8, Q7L3U6, Q9BZ39 | Gene names | FAM54A, DUFD1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM54A (DUF729 domain-containing protein 1). | |||||
|
FA54A_MOUSE
|
||||||
| NC score | 0.971031 (rank : 2) | θ value | 1.17728e-108 (rank : 2) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 29 | Shared Neighborhood Hits | 8 | |
| SwissProt Accessions | Q8VED8 | Gene names | Fam54a, Dufd1 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM54A (DUF729 domain-containing protein 1). | |||||
|
MTFR1_MOUSE
|
||||||
| NC score | 0.761785 (rank : 3) | θ value | 2.12685e-25 (rank : 3) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 38 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q99MB2, Q9CQZ4 | Gene names | Mtfr1, Kiaa0009 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitochondrial fission regulator 1. | |||||
|
MTFR1_HUMAN
|
||||||
| NC score | 0.747268 (rank : 4) | θ value | 3.07116e-24 (rank : 4) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 17 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q15390, Q6IB94, Q86XH5, Q8IVD7 | Gene names | MTFR1, CHPPR, KIAA0009 | |||
|
Domain Architecture |

|
|||||
| Description | Mitochondrial fission regulator 1 (Chondrocyte protein with a poly- proline region). | |||||
|
PRR11_MOUSE
|
||||||
| NC score | 0.082455 (rank : 5) | θ value | 0.813845 (rank : 6) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 36 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q8BHE0, Q3V3H1 | Gene names | Prr11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 11. | |||||
|
FA47B_HUMAN
|
||||||
| NC score | 0.070142 (rank : 6) | θ value | 0.00869519 (rank : 5) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 431 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q8NA70, Q5JQN5, Q6PIG3 | Gene names | FAM47B | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Protein FAM47B. | |||||
|
PRR11_HUMAN
|
||||||
| NC score | 0.061722 (rank : 7) | θ value | 5.27518 (rank : 14) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 27 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q96HE9, Q9NUZ7, Q9NXE9 | Gene names | PRR11 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Proline-rich protein 11. | |||||
|
TSH3_MOUSE
|
||||||
| NC score | 0.031719 (rank : 8) | θ value | 5.27518 (rank : 19) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 590 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8CGV9, Q5DTX4 | Gene names | Tshz3, Kiaa1474, Tsh3, Zfp537, Znf537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
TSH3_HUMAN
|
||||||
| NC score | 0.031638 (rank : 9) | θ value | 5.27518 (rank : 18) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 564 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q63HK5, Q9H0G6, Q9P254 | Gene names | TSHZ3, KIAA1474, TSH3, ZNF537 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Teashirt homolog 3 (Zinc finger protein 537). | |||||
|
CEBPE_HUMAN
|
||||||
| NC score | 0.026525 (rank : 10) | θ value | 1.81305 (rank : 7) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 236 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q15744, Q15745, Q8IYI2, Q99803 | Gene names | CEBPE | |||
|
Domain Architecture |

|
|||||
| Description | CCAAT/enhancer-binding protein epsilon (C/EBP epsilon). | |||||
|
SFRS4_MOUSE
|
||||||
| NC score | 0.024473 (rank : 11) | θ value | 1.81305 (rank : 9) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 406 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | Q8VE97, Q9JJC3 | Gene names | Sfrs4 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4. | |||||
|
SFRS4_HUMAN
|
||||||
| NC score | 0.024205 (rank : 12) | θ value | 1.81305 (rank : 8) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 384 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08170, Q5VXP1, Q9BUA4, Q9UEB5 | Gene names | SFRS4, SRP75 | |||
|
Domain Architecture |

|
|||||
| Description | Splicing factor, arginine/serine-rich 4 (Pre-mRNA-splicing factor SRP75) (SRP001LB). | |||||
|
GON4L_MOUSE
|
||||||
| NC score | 0.018657 (rank : 13) | θ value | 4.03905 (rank : 13) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 455 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9DB00, Q80TB4, Q91YI9 | Gene names | Gon4l, Gon4, KIAA1606 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | GON-4-like protein (GON-4 homolog). | |||||
|
RNF12_MOUSE
|
||||||
| NC score | 0.015674 (rank : 14) | θ value | 5.27518 (rank : 15) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 159 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q9WTV7 | Gene names | Rnf12, Rlim | |||
|
Domain Architecture |

|
|||||
| Description | RING finger protein 12 (LIM domain-interacting RING finger protein) (RING finger LIM domain-binding protein) (R-LIM). | |||||
|
SPRL1_HUMAN
|
||||||
| NC score | 0.014056 (rank : 15) | θ value | 6.88961 (rank : 21) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 231 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q14515, Q14800 | Gene names | SPARCL1 | |||
|
Domain Architecture |

|
|||||
| Description | SPARC-like protein 1 precursor (High endothelial venule protein) (Hevin) (MAST 9). | |||||
|
TLE3_MOUSE
|
||||||
| NC score | 0.013092 (rank : 16) | θ value | 5.27518 (rank : 17) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 378 | Shared Neighborhood Hits | 4 | |
| SwissProt Accessions | Q08122, Q923A4 | Gene names | Tle3, Esg | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG) (Grg-3). | |||||
|
TLE3_HUMAN
|
||||||
| NC score | 0.012998 (rank : 17) | θ value | 5.27518 (rank : 16) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 385 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | Q04726, Q8IVV6, Q8WVR2, Q9HCM5 | Gene names | TLE3, KIAA1547 | |||
|
Domain Architecture |

|
|||||
| Description | Transducin-like enhancer protein 3 (ESG3). | |||||
|
MAP2_MOUSE
|
||||||
| NC score | 0.012277 (rank : 18) | θ value | 8.99809 (rank : 23) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 342 | Shared Neighborhood Hits | 6 | |
| SwissProt Accessions | P20357 | Gene names | Map2, Mtap2 | |||
|
Domain Architecture |

|
|||||
| Description | Microtubule-associated protein 2 (MAP 2). | |||||
|
TAB2_HUMAN
|
||||||
| NC score | 0.012271 (rank : 19) | θ value | 8.99809 (rank : 24) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 169 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q9NYJ8, O94838, Q6I9W8, Q76N06, Q9UFP7 | Gene names | MAP3K7IP2, KIAA0733, TAB2 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | Mitogen-activated protein kinase kinase kinase 7-interacting protein 2 (TAK1-binding protein 2). | |||||
|
EP15_HUMAN
|
||||||
| NC score | 0.012260 (rank : 20) | θ value | 2.36792 (rank : 11) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 1021 | Shared Neighborhood Hits | 5 | |
| SwissProt Accessions | P42566 | Gene names | EPS15, AF1P | |||
|
Domain Architecture |

|
|||||
| Description | Epidermal growth factor receptor substrate 15 (Protein Eps15) (AF-1p protein). | |||||
|
2DMB_HUMAN
|
||||||
| NC score | 0.012187 (rank : 21) | θ value | 2.36792 (rank : 10) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 216 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | P28068, O77936, Q13012, Q29751, Q9XRX2 | Gene names | HLA-DMB, DMB, RING7 | |||
|
Domain Architecture |

|
|||||
| Description | HLA class II histocompatibility antigen, DM beta chain precursor (MHC class II antigen DMB). | |||||
|
CAC1G_HUMAN
|
||||||
| NC score | 0.012107 (rank : 22) | θ value | 3.0926 (rank : 12) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 587 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | O43497, O43498, O94770, Q9NYU4, Q9NYU5, Q9NYU6, Q9NYU7, Q9NYU8, Q9NYU9, Q9NYV0, Q9NYV1, Q9UHN9, Q9UHP0, Q9ULU6, Q9UNG7, Q9Y5T2, Q9Y5T3 | Gene names | CACNA1G, KIAA1123 | |||
|
Domain Architecture |

|
|||||
| Description | Voltage-dependent T-type calcium channel subunit alpha-1G (Voltage- gated calcium channel subunit alpha Cav3.1) (Cav3.1c) (NBR13). | |||||
|
ABCAD_MOUSE
|
||||||
| NC score | 0.007483 (rank : 23) | θ value | 6.88961 (rank : 20) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 150 | Shared Neighborhood Hits | 2 | |
| SwissProt Accessions | Q5SSE9, Q80T20, Q8BHZ2, Q8CB91 | Gene names | Abca13 | |||
|
Domain Architecture |
No domain graphic available |
|||||
| Description | ATP-binding cassette sub-family A member 13. | |||||
|
CLC4F_MOUSE
|
||||||
| NC score | 0.005402 (rank : 24) | θ value | 8.99809 (rank : 22) | |||
| Query Neighborhood Hits | 24 | Target Neighborhood Hits | 572 | Shared Neighborhood Hits | 3 | |
| SwissProt Accessions | P70194, Q8BLZ8 | Gene names | Clec4f, Clecsf13, Kclr | |||
|
Domain Architecture |

|
|||||
| Description | C-type lectin domain family 4 member F (C-type lectin superfamily member 13) (C-type lectin 13) (Kupffer cell receptor). | |||||